8TPG
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | (3R)-N-(4-tert-butylphenyl)-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-3-hydroxybutanamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
8EOZ
 
 | |
6BTI
 
 | | Crystal structure of human cellular retinol binding protein 2 (CRBP2) in complex with N-arachidonoylethanolamine (AEA) | | Descriptor: | (5Z,8Z,11Z,14Z)-N-(2-hydroxyethyl)icosa-5,8,11,14-tetraenamide, DI(HYDROXYETHYL)ETHER, Retinol-binding protein 2 | | Authors: | Silvaroli, J.A, Blaner, W.S, Lodowski, D.T, Golczak, M. | | Deposit date: | 2017-12-06 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Retinol-binding protein 2 (RBP2) binds monoacylglycerols and modulates gut endocrine signaling and body weight.
Sci Adv, 6, 2020
|
|
6VQL
 
 | |
8TPC
 
 | | Synthesis, X-Ray Crystallographic and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease | | Descriptor: | N-[(1R)-2-(benzylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-[4-(2-chloroacetamido)phenyl]furan-2-carboxamide, Non-structural protein 7 | | Authors: | Chua, T.K, Song, Y. | | Deposit date: | 2023-08-04 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Design, Synthesis, X-ray Crystallography, and Biological Activities of Covalent, Non-Peptidic Inhibitors of SARS-CoV-2 Main Protease.
Acs Infect Dis., 10, 2024
|
|
6BUG
 
 | |
8ERW
 
 | | Precisely patterned nanofibers made from extendable protein multiplexes | | Descriptor: | C2HR4_8r | | Authors: | Bick, M.J, Parmeggiani, F, Bethel, N.P, Sankaran, B, Baker, D. | | Deposit date: | 2022-10-12 | | Release date: | 2023-08-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Precisely patterned nanofibres made from extendable protein multiplexes.
Nat.Chem., 15, 2023
|
|
2M8T
 
 | |
5FOG
 
 | | Crystal structure of hte Cryptosporidium muris cytosolic leucyl-tRNA synthetase editing domain complex with a post-transfer editing analogue of norvaline (Nv2AA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-(L-NORVALYL)AMINO-2'-DEOXYADENOSINE, LEUCYL-TRNA SYNTHETASE, ... | | Authors: | Palencia, A, Liu, R.J, Lukarska, M, Gut, J, Bougdour, A, Touquet, B, Wang, E.D, Alley, M.R.K, Rosenthal, P.J, Hakimi, M.A, Cusack, S. | | Deposit date: | 2015-11-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cryptosporidium and Toxoplasma Parasites are Inhibited by a Benzoxaborole Targeting Leucyl-tRNA Synthetase.
Antimicrob.Agents Chemother., 60, 2016
|
|
7E58
 
 | | interferon-inducible anti-viral protein 2 | | Descriptor: | Guanylate-binding protein 2 | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E5A
 
 | | interferon-inducible anti-viral protein R356A | | Descriptor: | ALUMINUM FLUORIDE, GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 5, ... | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7E59
 
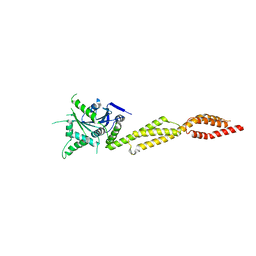 | | interferon-inducible anti-viral protein truncated | | Descriptor: | Guanylate-binding protein 5 | | Authors: | Cui, W, Wang, W, Chen, C, Slater, B, Xiong, Y, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for GTP-induced dimerization and antiviral function of guanylate-binding proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6DWY
 
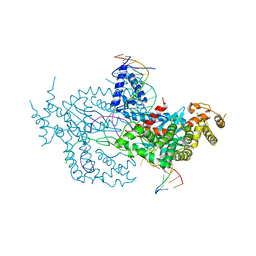 | | Hermes transposase deletion dimer complex with (C/G) DNA and Ca2+ | | Descriptor: | CALCIUM ION, DNA (26-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Dyda, F, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
5EDX
 
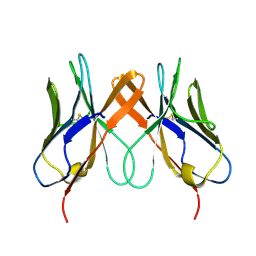 | | Crystal structure of swine CD8aa homodimer | | Descriptor: | CD8 alpha antigen | | Authors: | Liu, Y.J, Qi, J.X, Zhang, N.Z, Xia, C. | | Deposit date: | 2015-10-22 | | Release date: | 2016-09-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
6DWW
 
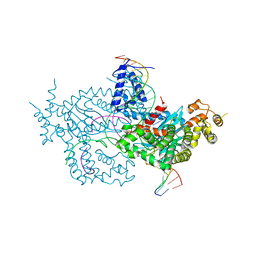 | | Hermes transposase deletion dimer complex with (A/T) DNA and Mn2+ | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
6DX0
 
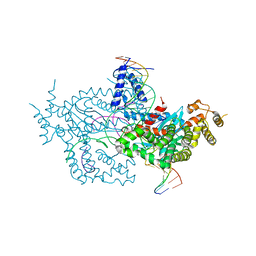 | | Hermes transposase deletion dimer complex with (A/T) DNA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
5FGD
 
 | |
6UD9
 
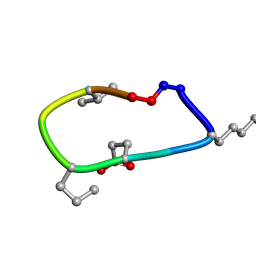 | | S2 symmetric peptide design number 2, Morticia | | Descriptor: | S2-2, Morticia | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-19 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UCX
 
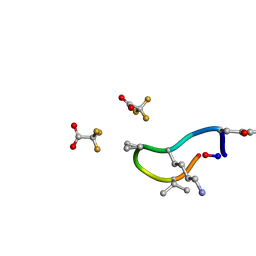 | | S2 symmetric peptide design number 1, Wednesday | | Descriptor: | S2-1, Wednesday, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-18 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
6UDZ
 
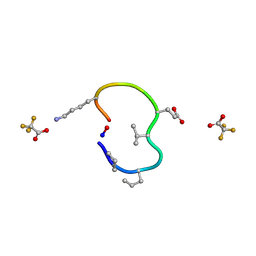 | | S2 symmetric peptide design number 4 crystal form 1, Pugsley | | Descriptor: | S2-4, Pusgley crystal form 1, trifluoroacetic acid | | Authors: | Mulligan, V.K, Kang, C.S, Antselovich, I, Sawaya, M.R, Yeates, T.O, Baker, D. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Computational design of mixed chirality peptide macrocycles with internal symmetry.
Protein Sci., 29, 2020
|
|
7WZ7
 
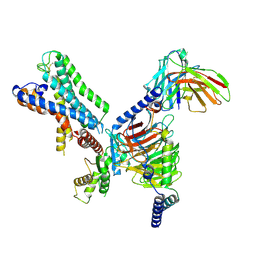 | | GPR110/G12 complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7WXU
 
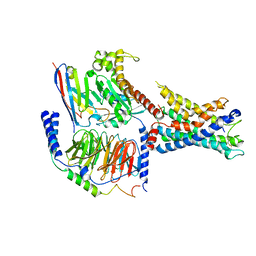 | | GPR110/Gq complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
6DZR
 
 | | Crystal structure of h38C2 K99R mutation | | Descriptor: | CITRATE ANION, SULFATE ION, h38c2 heavy chain, ... | | Authors: | Park, H, Rader, C. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-Selective Antibody Functionalization via Orthogonally Reactive Arginine and Lysine Residues.
Cell Chem Biol, 26, 2019
|
|
7WY0
 
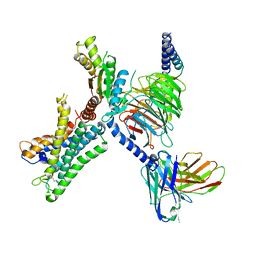 | | GPR110/G13 complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Engineered G alpha 13 subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
7WXW
 
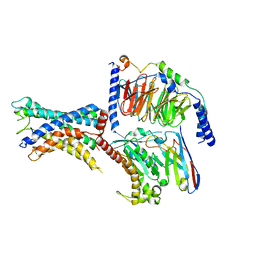 | | GPR110/Gs complex | | Descriptor: | Adhesion G-protein coupled receptor F1, Engineered mini Galpha-s subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2022-02-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis of adhesion GPCR GPR110 activation by stalk peptide and G-proteins coupling.
Nat Commun, 13, 2022
|
|
