7JGU
 
 | | Structure of FN3tt mut | | Descriptor: | Fibronectin type-III domain-containing protein, PENTAETHYLENE GLYCOL | | Authors: | Luo, J, Boucher, L.E. | | Deposit date: | 2020-07-19 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Surface salt bridges contribute to the extreme thermal stability of an FN3-like domain from a thermophilic bacterium.
Proteins, 90, 2022
|
|
7XZ4
 
 | |
7Y56
 
 | |
7XUP
 
 | | Crystal structure of TPe3.0 | | Descriptor: | Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
7XUQ
 
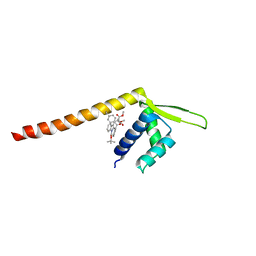 | | Crystal structure of Tpe3.0 complexed with N-Boc-3-alkenylindole | | Descriptor: | Transcriptional regulator, PadR-like family, dimethyl 2-[[2-methyl-1-[(2-methylpropan-2-yl)oxycarbonyl]indol-3-yl]methyl]-2-prop-2-enyl-propanedioate | | Authors: | Chen, X, Qian, J.Y, Sun, N.N, Zhong, F.R, Wu, Y.Z. | | Deposit date: | 2022-05-19 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enantioselective [2+2]-cycloadditions with triplet photoenzymes.
Nature, 611, 2022
|
|
1XNN
 
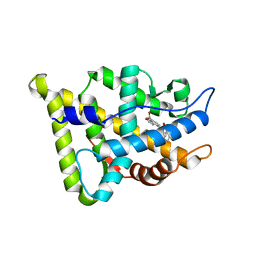 | | CRYSTAL STRUCTURE OF THE RAT ANDROGEN RECEPTOR LIGAND BINDING DOMAIN T877A MUTANT COMPLEX WITH (3A-ALPHA-,4-ALPHA 7-ALPHA-,7A-ALPHA-)-3A,4,7,7A-TETRAHYDRO-2-(4-NITRO-1-NAPHTHALENYL)-4,7-ETHANO-1H-ISOINDOLE-1,3(2H)-DIONE. | | Descriptor: | Androgen receptor, REL-(3AR,4S,7R,7AS)-3A,4,7,7A-TETRAHYDRO-2-(4-NITRO-1-NAPHTHALENYL)-4,7-ETHANO-1H-ISOINDOLE-1,3(2H)-DIONE | | Authors: | Sack, J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure based approach to the design of bicyclic-1H-isoindole-1,3(2H)-dione based androgen receptor antagonists.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
7WH9
 
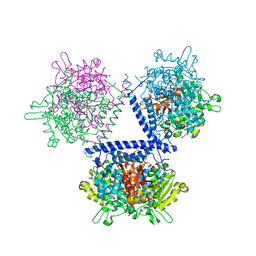 | | holo structure of emodin 1-OH O-methyltransferase complex with emodin and S-Adenosyl-L-homocysteine | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, O-methyltransferase gedA, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Liang, Y.J, Lu, X.F, Qi, F.F, Xue, Y.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Characterization and Structural Analysis of Emodin- O -Methyltransferase from Aspergillus terreus.
J.Agric.Food Chem., 70, 2022
|
|
3UQP
 
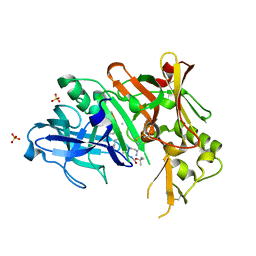 | | Crystal structure of Bace1 with its inhibitor | | Descriptor: | Beta-secretase 1, METHYL (2R)-1-[(6S,9S,12S,13S,17S,20S,23R)-9-(3-AMINO-3-OXOPROPYL)-12,23-DIBENZYL-13-HYDROXY-2,2,8,20,22-PENTAMETHYL-17-(2-METHYLPROPYL)-4,7,10,15,18,21,24-HEPTAOXO-6-(PROPAN-2-YL)-3-OXA-5,8,11,16,19,22-HEXAAZATETRACOSAN-24-YL]PYRROLIDINE-2-CARBOXYLATE, SULFATE ION | | Authors: | Chen, T.T, Chen, W.Y, Li, L, Xu, Y.C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Cyanobacterial Peptides as a Prototype for the Design of Potent beta-Secretase Inhibitors and the Development of Selective Chemical Probes for Other Aspartic Proteases
J.Med.Chem., 55, 2012
|
|
7KPY
 
 | | Crystal structure of CBP bromodomain liganded with UMB298 (compound 23) | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(3-chloranyl-4-methoxy-phenyl)ethyl]-~{N}-cyclohexyl-7-(3,5-dimethyl-1,2-oxazol-4-yl)imidazo[1,2-a]pyridin-3-amine, Histone acetyltransferase | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of Dimethylisoxazole-Attached Imidazo[1,2- a ]pyridines as Potent and Selective CBP/P300 Inhibitors.
J.Med.Chem., 64, 2021
|
|
4K91
 
 | | Crystal structure of Penicillin-Binding Protein 5 (PBP5) from Pseudomonas aeruginosa in apo state | | Descriptor: | D-ala-D-ala-carboxypeptidase, SUCCINIC ACID | | Authors: | Smith, J, Toth, M, Vakulenko, S, Mobashery, S, Chen, Y. | | Deposit date: | 2013-04-19 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural analysis of the role of Pseudomonas aeruginosa penicillin-binding protein 5 in beta-lactam resistance.
Antimicrob.Agents Chemother., 57, 2013
|
|
1YIN
 
 | |
4O6Z
 
 | | Crystal structure of serine hydroxymethyltransferase with covalently bound PLP Schiff-base from Plasmodium falciparum | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2013-12-24 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The structure of Plasmodium falciparum serine hydroxymethyltransferase reveals a novel redox switch that regulates its activities.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1YIM
 
 | |
3WXA
 
 | | X-ray crystal structural analysis of the complex between ALG-2 and Sec31A peptide | | Descriptor: | Programmed cell death protein 6, Protein transport protein Sec31A, ZINC ION | | Authors: | Takahashi, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-03-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Analysis of the Complex between Penta-EF-Hand ALG-2 Protein and Sec31A Peptide Reveals a Novel Target Recognition Mechanism of ALG-2
Int J Mol Sci, 16, 2015
|
|
2IOG
 
 | |
2IOK
 
 | |
7WVD
 
 | | Crystal structure of H14 complexed with SIA28 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SIA28, ... | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7WVI
 
 | | Crystal structure of SIA28 | | Descriptor: | Heavy chain of SIA28, Light chain of SIA28 | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
7WVG
 
 | | Crystal structure of H18 complexed with SIA28 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SIA28, ... | | Authors: | Chen, Y, Qi, J, Gao, G.F. | | Deposit date: | 2022-02-10 | | Release date: | 2023-01-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a human broadly neutralizing influenza A hemagglutinin stem-specific antibody including H17/18 subtypes.
Nat Commun, 13, 2022
|
|
1S6Q
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R147681 | | Descriptor: | 4-[4-(2,4,6-TRIMETHYL-PHENYLAMINO)-PYRIMIDIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
6BTM
 
 | | Structure of Alternative Complex III from Flavobacterium johnsoniae (Wild Type) | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl ditetradecanoate, Alternative Complex III subunit A, Alternative Complex III subunit B, ... | | Authors: | Sun, C, Benlekbir, S, Venkatakrishnan, P, Yuhang, W, Tajkhorshid, E, Rubinstein, J.L, Gennis, R.B. | | Deposit date: | 2017-12-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the alternative complex III in a supercomplex with cytochrome oxidase.
Nature, 557, 2018
|
|
1S6P
 
 | | CRYSTAL STRUCTURE OF HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R100943 | | Descriptor: | 1-(4-CYANO-PHENYL)-3-[2-(2,6-DICHLORO-PHENYL)-1-IMINO-ETHYL]-THIOUREA, MAGNESIUM ION, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-01-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1S9E
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R129385 | | Descriptor: | 4-[4-AMINO-6-(2,6-DICHLORO-PHENOXY)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase], POL polyprotein [Contains:Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | Descriptor: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | Authors: | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | Deposit date: | 2004-02-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
1SCY
 
 | |
