3MTC
 
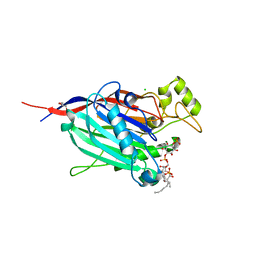 | | Crystal Structure of INPP5B in complex with phosphatidylinositol 4-phosphate | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dioctanoate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, van der Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for phosphoinositide substrate recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases
Structure, 22, 2014
|
|
3SOG
 
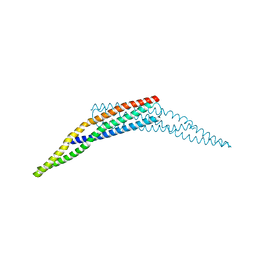 | | Crystal structure of the BAR domain of human Amphiphysin, isoform 1 | | Descriptor: | 1,2-ETHANEDIOL, Amphiphysin, POTASSIUM ION | | Authors: | Allerston, C.K, Krojer, T, Chaikuad, A, Cooper, C.D.O, Berridge, G, Savitsky, P, Vollmar, M, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: |
|
|
3SOM
 
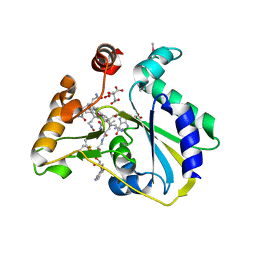 | | crystal structure of human MMACHC | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-ETHANETHIOL, 5'-DEOXYADENOSINE, ... | | Authors: | Krojer, T, Froese, D.S, von Delft, F, Muniz, J.R, Gileadi, C, Vollmar, M, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Gravel, R.A, Yue, W.W, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-30 | | Release date: | 2011-07-27 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of MMACHC reveals an arginine-rich pocket and a domain-swapped dimer for its B12 processing function.
Biochemistry, 51, 2012
|
|
2I7P
 
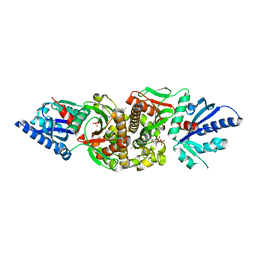 | | Crystal structure of human PANK3 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, Pantothenate kinase 3 | | Authors: | Hong, B.S, Wang, L, Shen, L, Tempel, W, Loppnau, P, Finerty, P, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-31 | | Release date: | 2006-12-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of human pantothenate kinases. Insights into allosteric regulation and mutations linked to a neurodegeneration disorder.
J.Biol.Chem., 282, 2007
|
|
2I75
 
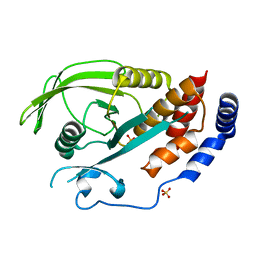 | | Crystal Structure of Human Protein Tyrosine Phosphatase N4 (PTPN4) | | Descriptor: | SULFATE ION, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Ugochukwu, E, Barr, A, Savitsky, P, Burgess, N, Das, S, Turnbull, A, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
3Q4S
 
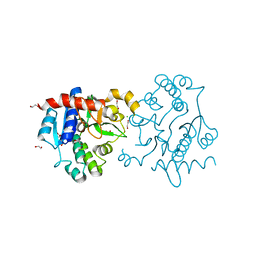 | | Crystal Structure of Human Glycogenin-1 (GYG1), apo form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glycogenin-1 | | Authors: | Chaikuad, A, Froese, D.S, Yue, W.W, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-24 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2GTP
 
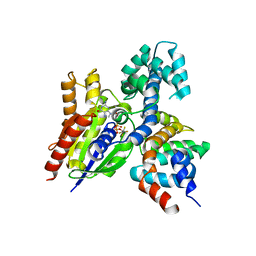 | | Crystal structure of the heterodimeric complex of human RGS1 and activated Gi alpha 1 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Soundararajan, M, Turnbull, A.P, Ugochukwu, E, Gorrec, F, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural diversity in the RGS domain and its interaction with heterotrimeric G protein alpha-subunits.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3N50
 
 | | Human Early B-cell factor 3 (EBF3) IPT/TIG and HLHLH domains | | Descriptor: | Transcription factor COE3 | | Authors: | Lehtio, L, Siponen, M.I, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schuler, H, Schutz, P, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Wisniewska, M, Berglund, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-24 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Structural Determination of Functional Domains in Early B-cell Factor (EBF) Family of Transcription Factors Reveals Similarities to Rel DNA-binding Proteins and a Novel Dimerization Motif.
J.Biol.Chem., 285, 2010
|
|
2I3Y
 
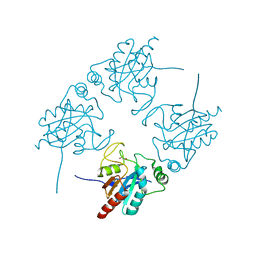 | | Crystal structure of human glutathione peroxidase 5 | | Descriptor: | 1,2-ETHANEDIOL, Epididymal secretory glutathione peroxidase | | Authors: | Kavanagh, K.L, Johansson, C, Rojkova, A, Umeano, C, Bunkoczi, G, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-21 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human glutathione peroxidase 5
To be published
|
|
2OQ6
 
 | | Crystal structure of JMJD2A complexed with histone H3 peptide trimethylated at Lys9 | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Pilka, E.S, Ng, S.S, Kavanagh, K.L, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2OO1
 
 | | Crystal structure of the Bromo domain 2 of human Bromodomain containing protein 3 (BRD3) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Bromodomain-containing protein 3, ... | | Authors: | Filippakopoulos, P, Bullock, A, Papagrigoriou, E, Keates, T, Cooper, C, Smee, C, Ugochukwu, E, Debreczeni, J, von Delft, F, Arrowsmith, C, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-25 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2OQ7
 
 | | The crystal structure of JMJD2A complexed with Ni and N-oxalylglycine | | Descriptor: | JmjC domain-containing histone demethylation protein 3A, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Kavanagh, K.L, Ng, S.S, Pilka, E, McDonough, M.A, Savitsky, P, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Sundstrom, M, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-31 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of histone demethylase JMJD2A reveal basis for substrate specificity.
Nature, 448, 2007
|
|
2QGX
 
 | | Ubiquitin-conjugating enzyme E2Q | | Descriptor: | Ubiquitin-conjugating enzyme E2 Q1 | | Authors: | Neculai, D, Avvakumov, G.V, Xue, S, Walker, J.R, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Sicheri, F, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
3QVB
 
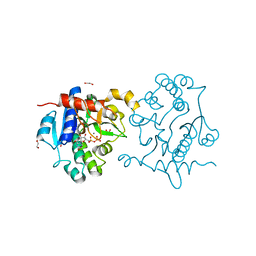 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese and UDP | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Yue, W.W, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-25 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U2U
 
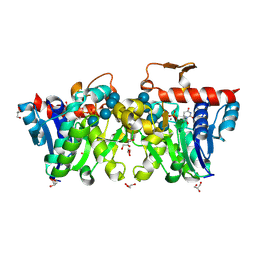 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese, UDP and maltotetraose | | Descriptor: | GLYCEROL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3U2T
 
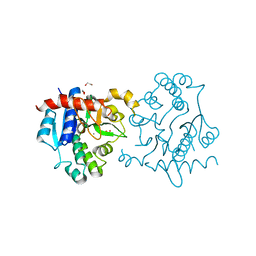 | | Crystal Structure of Human Glycogenin-1 (GYG1) complexed with manganese | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION | | Authors: | Chaikuad, A, Froese, D.S, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, U, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2JFD
 
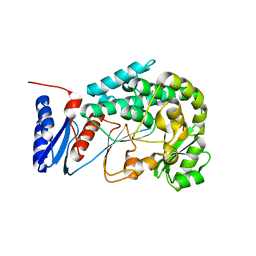 | | Structure of the MAT domain of human FAS | | Descriptor: | FATTY ACID SYNTHASE | | Authors: | Bunkoczi, G, Kavanagh, K, Hozjan, V, Rojkova, A, Wu, X, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Smith, S, Oppermann, U. | | Deposit date: | 2007-01-31 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for different specificities of acyltransferases associated with the human cytosolic and mitochondrial fatty acid synthases.
Chem. Biol., 16, 2009
|
|
3UVJ
 
 | | Crystal structure of the catalytic domain of the heterodimeric human soluble guanylate cyclase 1. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Guanylate cyclase soluble subunit alpha-3, ... | | Authors: | Allerston, C.K, Berridge, G, Chalk, R, Cooper, C.D.O, Savitsky, P, Vollmar, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-11-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of the catalytic domain of human soluble guanylate cyclase.
Plos One, 8, 2013
|
|
3V2B
 
 | | Human poly(adp-ribose) polymerase 15 (ARTD7, BAL3), macro domain 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 15, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Moche, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kotenyova, T, Kotzcsh, A, Kraulis, P, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A.K, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van den berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-12 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
3VFQ
 
 | | Human PARP14 (ARTD8, BAL2) - macro domains 1 and 2 in complex with adenosine-5-diphosphoribose | | Descriptor: | Poly [ADP-ribose] polymerase 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-10 | | Release date: | 2013-03-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recognition of Mono-ADP-Ribosylated ARTD10 Substrates by ARTD8 Macrodomains.
Structure, 21, 2013
|
|
2Y7F
 
 | |
2OC3
 
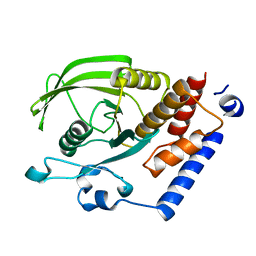 | | Crystal Structure of the Catalytic Domain of Human Protein Tyrosine Phosphatase non-receptor Type 18 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Ugochukwu, E, Barr, A, Alfano, I, Gorrec, F, Umeano, C, Savitsky, P, Sobott, F, Eswaran, J, Papagrigoriou, E, Debreczeni, J.E, Turnbull, A, Bunkoczi, G, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-12-20 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
5V38
 
 | |
2PQF
 
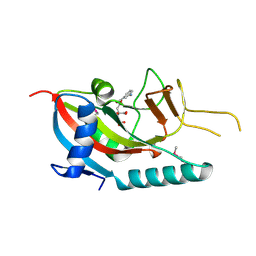 | | Human Poly(ADP-Ribose) Polymerase 12, Catalytic fragment in complex with an inhibitor 3-Aminobenzoic acid | | Descriptor: | 3-AMINOBENZOIC ACID, CITRIC ACID, Poly [ADP-ribose] polymerase 12 | | Authors: | Karlberg, T, Lehtio, L, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hogbom, M, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-02 | | Release date: | 2007-05-15 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
2PSO
 
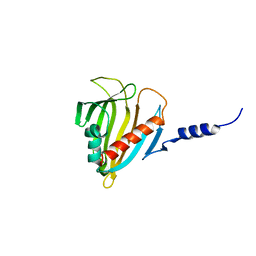 | | Human StarD13 (DLC2) lipid transfer and protein localization domain | | Descriptor: | StAR-related lipid transfer protein 13 | | Authors: | Lehtio, L, Busam, R, Arrowsmith, C.H, Berglund, H, Collins, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg-Schiavone, L, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Moche, M, Nordlund, P, Nyman, T, Ogg, D, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van den Berg, S, Weigelt, J, Welin, M, Persson, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-07 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
