7X46
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X47
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X3C
 
 | | Cryo-EM structure of Coxsackievirus B1 muture virion in complex with nAbs 8A10 and 5F5 (CVB1-M:8A10:5F5) | | Descriptor: | 5F5 heavy chain, 5F5 light chain, 8A10 heavy chain, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4M
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion in complex with nAb 8A10 (classified from CVB1 mature virion in complex with 8A10, 2E6 and 9A3) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, Capsid protein VP4, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7Y97
 
 | | Crystal structure of CYP109B4 from Bacillus Sonorensis | | Descriptor: | Cytochrome P450 monooxygenase YjiB, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shen, P.P, Huang, J.-W, Li, X, Liu, W.D, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family
Acs Catalysis, 13, 2023
|
|
7Y9O
 
 | | Crystal structure of a CYP109B4 variant from Bacillus sonorensis | | Descriptor: | CALCIUM ION, Cytochrome P450 monooxygenase YjiB, IMIDAZOLE, ... | | Authors: | Shen, P.P, Huang, J.-W, Li, X, Liu, W.D, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family
Acs Catalysis, 13, 2023
|
|
3RX4
 
 | | Crystal Structure of Human Aldose Reductase complexed with Sulindac Sulfide | | Descriptor: | 2-[(3Z)-6-fluoranyl-2-methyl-3-[(4-methylsulfanylphenyl)methylidene]inden-1-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Chen, J, Luo, H, Hu, X. | | Deposit date: | 2011-05-10 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
3RX2
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Sulindac Sulfone | | Descriptor: | 2-[(3Z)-6-fluoranyl-2-methyl-3-[(4-methylsulfonylphenyl)methylidene]inden-1-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, X, Chen, J, Luo, H, Hu, X. | | Deposit date: | 2011-05-10 | | Release date: | 2011-11-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase
Febs Lett., 586, 2012
|
|
3S3G
 
 | | Crystal Structure of Human Aldose Reductase Complexed with Tolmetin | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tolmetin | | Authors: | Zheng, X, Chen, J, Luo, H, Hu, X. | | Deposit date: | 2011-05-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The molecular basis for inhibition of sulindac and its metabolites towards human aldose reductase.
Febs Lett., 586, 2012
|
|
7YUF
 
 | | apo human SPNS2 | | Descriptor: | NbFab H-chain, NbFab L-chain, Sphingosine-1-phosphate transporter SPNS2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YUD
 
 | | FTY720p-bound human SPNS2 | | Descriptor: | (2~{S})-2-azanyl-4-(4-octylphenyl)-2-[[oxidanyl-bis(oxidanylidene)-$l^{6}-phosphanyl]oxymethyl]butan-1-ol, NbFab L-chain, NbFab-H-chain, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2022-08-17 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis of Sphingosine-1-phosphate transport via human SPNS2.
Cell Res., 34, 2024
|
|
7YMD
 
 | | Cryo-EM structure of Nse1/3/4 | | Descriptor: | Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, Non-structural maintenance of chromosomes element 1 | | Authors: | Qian, L, Jun, Z, Zhenguo, C, Wang, L. | | Deposit date: | 2022-07-28 | | Release date: | 2024-01-31 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.176 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
6PSH
 
 | |
6QNS
 
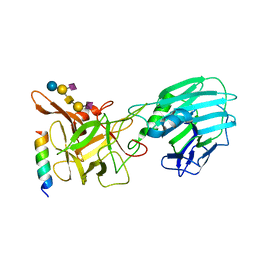 | | Crystal structure of the binding domain of Botulinum Neurotoxin type B mutant I1248W/V1249W in complex with human synaptotagmin 1 and GD1a receptors | | Descriptor: | Botulinum neurotoxin type B, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-[N-acetyl-alpha-neuraminic acid-(2-3)]beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Synaptotagmin-1 | | Authors: | Masuyer, G, Yin, L, Zhang, S, Miyashita, S.I, Dong, M, Stenmark, P. | | Deposit date: | 2019-02-12 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization of a membrane binding loop leads to engineering botulinum neurotoxin B with improved therapeutic efficacy.
Plos Biol., 18, 2020
|
|
7TAD
 
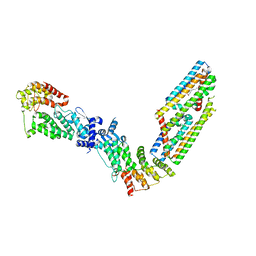 | | CryoEM structure of the (NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6WJ5
 
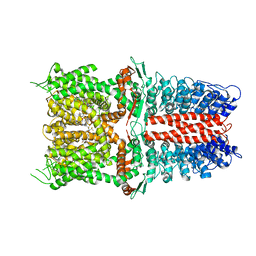 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
7TAC
 
 | | Cryo-EM structure of the (TGA3)2-(NPR1)2-(TGA3)2 complex | | Descriptor: | PALMITIC ACID, Regulatory protein NPR1, Transcription factor TGA3, ... | | Authors: | Wu, Q, Zhou, Y, Bartesaghi, A, Dong, X, Zhou, P. | | Deposit date: | 2021-12-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
6PXE
 
 | |
6PSK
 
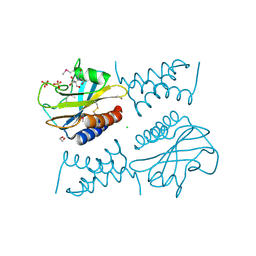 | | Crystal structure of the complex between periplasmic domains of antiholin RI and holin T from T4 phage, in P6522 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Antiholin, ... | | Authors: | Kuznetsov, V.B, Krieger, I.V, Sacchettini, J.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of T4 Phage Lysis Control: DNA as the Signal for Lysis Inhibition.
J.Mol.Biol., 432, 2020
|
|
6PX4
 
 | |
8RZ3
 
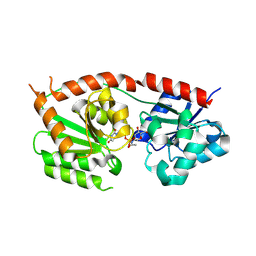 | | Structures of Se- glycosyltransferase SenB from Variovorax paradoxus | | Descriptor: | TIGR04348 family glycosyltransferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Ma, Y.Y, Gao, Y, Xu, S.H. | | Deposit date: | 2024-02-12 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of SenB and SenA enzymes from Variovorax paradoxus provide insights into carbon-selenium bond formation in selenoneine biosynthesis.
Heliyon, 10, 2024
|
|
8RYZ
 
 | | Structures of selenoneine synthase SenA from Variovorax paradoxus | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, IMIDAZOLE, ... | | Authors: | Ma, Y.Y, Gao, Y, Xu, S.H. | | Deposit date: | 2024-02-11 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structures of SenB and SenA enzymes from Variovorax paradoxus provide insights into carbon-selenium bond formation in selenoneine biosynthesis.
Heliyon, 10, 2024
|
|
8SBG
 
 | |
8SIJ
 
 | | Crystal structure of F. varium tryptophanase | | Descriptor: | CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE, Tryptophanase 1, ... | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
8SL7
 
 | | Butyricicoccus sp. BIOML-A1 tryptophanase complex with (3S) ALG-05 | | Descriptor: | (E)-3-[(3S)-3-chloro-2-oxo-2,3-dihydro-1H-indol-3-yl]-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-alanine, Tryptophanase | | Authors: | Graboski, A.L, Redinbo, M.R. | | Deposit date: | 2023-04-21 | | Release date: | 2023-08-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Mechanism-based inhibition of gut microbial tryptophanases reduces serum indoxyl sulfate.
Cell Chem Biol, 30, 2023
|
|
