6AKL
 
 | | Crystal structure of Striatin3 in complex with SIKE1 Coiled-coil domain | | Descriptor: | Striatin-3, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
3WL5
 
 | | Crystal structure of pOPH S172C | | Descriptor: | CITRIC ACID, Oxidized polyvinyl alcohol hydrolase | | Authors: | Yang, Y, Ko, T.P, Li, J.H, Liu, L, Huang, C.H, Chan, H.C, Ren, F.F, Jia, D.X, Wang, A.H.-J, Guo, R.T, Chen, J, Du, G.C. | | Deposit date: | 2013-11-07 | | Release date: | 2014-09-24 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into enzymatic degradation of oxidized polyvinyl alcohol
Chembiochem, 15, 2014
|
|
4CZ1
 
 | | Crystal structure of kynurenine formamidase from Bacillus anthracis complexed with 2-aminoacetophenone. | | Descriptor: | 2-aminoacetophenone, KYNURENINE FORMAMIDASE, MAGNESIUM ION, ... | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-04-16 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
5O8K
 
 | |
6H9X
 
 | | Klebsiella pneumoniae Seryl-tRNA Synthetase in Complex with the Intermediate Analog 5'-O-(N-(L-Seryl)-Sulfamoyl)Adenosine | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, CALCIUM ION, ... | | Authors: | Pang, L, De Graef, S, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2018-08-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acylated sulfonamide adenosines as potent inhibitors of the adenylate-forming enzyme superfamily.
Eur.J.Med.Chem., 174, 2019
|
|
4CNG
 
 | | Crystal structure of Sulfolobus acidocaldarius TrmJ in complex with S-adenosyl-L-Homocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, SPOU RRNA METHYLASE | | Authors: | Van Laer, B, Somme, J, Roovers, M, Steyaert, J, Droogmans, L, Versees, W. | | Deposit date: | 2014-01-22 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Characterization of Two Homologous 2'-O-Methyltransferases Showing Different Specificities for Their tRNA Substrates.
RNA, 20, 2014
|
|
5O8Z
 
 | | Conformational dynamism for DNA interaction in Salmonella typhimurium RcsB response regulator. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Transcriptional regulatory protein RcsB | | Authors: | Casino, P, Marina, A, Miguel-Romero, L, Huesa, J. | | Deposit date: | 2017-06-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conformational dynamism for DNA interaction in the Salmonella RcsB response regulator.
Nucleic Acids Res., 46, 2018
|
|
6HTU
 
 | | Structure of hStau1 dsRBD3-4 in complex with ARF1 RNA | | Descriptor: | Double-stranded RNA-binding protein Staufen homolog 1, RNA (19-MER) | | Authors: | Emmerich, C, Lazzaretti, D, Bandholz-Cajamarca, L, Bono, F. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | The crystal structure of Staufen1 in complex with a physiological RNA sheds light on substrate selectivity.
Life Sci Alliance, 1, 2018
|
|
5NXY
 
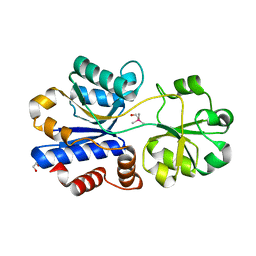 | | Crystal structure of OpuAC from B. subtilis in complex with Arsenobetaine | | Descriptor: | 1,2-ETHANEDIOL, 2-(trimethyl-lambda~5~-arsanyl)ethanol, Osmotically activated L-carnitine/choline ABC transporter substrate-binding protein OpuCC | | Authors: | Hofmann, T, Bremer, E, Schmitt, L, Smits, S. | | Deposit date: | 2017-05-11 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Arsenobetaine: an ecophysiologically important organoarsenical confers cytoprotection against osmotic stress and growth temperature extremes.
Environ. Microbiol., 20, 2018
|
|
1GAV
 
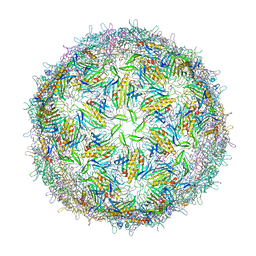 | | BACTERIOPHAGE GA PROTEIN CAPSID | | Descriptor: | BACTERIOPHAGE GA PROTEIN CAPSID | | Authors: | Tars, K, Bundule, M, Liljas, L. | | Deposit date: | 1997-01-28 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The crystal structure of bacteriophage GA and a comparison of bacteriophages belonging to the major groups of Escherichia coli leviviruses.
J.Mol.Biol., 271, 1997
|
|
8TL1
 
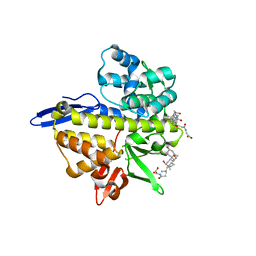 | |
1MG6
 
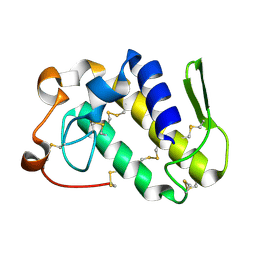 | |
5OBV
 
 | | Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with ADP and Pi. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5OC4
 
 | | Crystal structure of human tRNA-dihydrouridine(20) synthase dsRBD R361A-R362A mutant | | Descriptor: | CACODYLATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Bou-nader, C, Pecqueur, L, Hamdane, D. | | Deposit date: | 2017-06-29 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular basis for transfer RNA recognition by the double-stranded RNA-binding domain of human dihydrouridine synthase 2.
Nucleic Acids Res., 47, 2019
|
|
4D1S
 
 | | Pyrrole-3-carboxamides as potent and selective JAK2 inhibitors | | Descriptor: | 2-(5-chloro-2-methylphenyl)-1-methyl-5-(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)-1H-pyrrole-3-carboxamide, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Bertrand, J, Canevari, G, Fasolini, M, Brasca, M.G, Nesi, M, Avanzi, N, Ballinari, D, Bandiera, T, Bindi, S, Carenzi, D, Casero, D, Ceriani, L, Ciomei, M, Cirla, A, Colombo, M, Cribioli, S, Cristiani, C, Della Vedova, F, Fachin, G, Felder, E.R, Galvani, A, Isacchi, A, Mirizzi, D, Motto, I, Panzeri, A, Pesenti, E, Vianello, P, Gnocchi, P, Donati, D. | | Deposit date: | 2014-05-05 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Pyrrole-3-Carboxamides as Potent and Selective Jak2 Inhibitors.
Bioorg.Med.Chem., 22, 2014
|
|
3WQK
 
 | | Crystal structure of Rv3378c with PO4 | | Descriptor: | Diterpene synthase, PHOSPHATE ION | | Authors: | Chan, H.C, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Zheng, Y, Bogue, S, Nakano, C, Hoshino, T, Zhang, L, Lv, P, Liu, W, Crick, D.C, Liang, P.H, Wang, A.H, Oldfield, E, Guo, R.T. | | Deposit date: | 2014-01-28 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and inhibition of tuberculosinol synthase and decaprenyl diphosphate synthase from Mycobacterium tuberculosis.
J.Am.Chem.Soc., 136, 2014
|
|
6A32
 
 | |
8TKA
 
 | |
8TL8
 
 | | Structure of Orthoreovirus RNA Chaperone SigmaNS R6A mutant in complex with bile acid | | Descriptor: | GLYCOCHOLIC ACID, Protein sigma-NS | | Authors: | Prasad, B.V.V, Zhao, B, Hu, L, Neetu, N. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of orthoreovirus RNA chaperone sigma NS, a component of viral replication factories.
Nat Commun, 15, 2024
|
|
1FVS
 
 | | SOLUTION STRUCTURE OF THE YEAST COPPER TRANSPORTER DOMAIN CCC2A IN THE APO AND CU(I) LOAD STATES | | Descriptor: | COPPER (II) ION, COPPER-TRANSPORTING ATPASE | | Authors: | Banci, L, Bertini, I, Ciofi Baffoni, S, Huffman, D.L, O'Halloran, T.V. | | Deposit date: | 2000-09-20 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the yeast copper transporter domain Ccc2a in the apo and Cu(I)-loaded states.
J.Biol.Chem., 276, 2001
|
|
6HLR
 
 | | Yeast RNA polymerase I elongation complex bound to nucleotide analog GMPCPP (core focused) | | Descriptor: | DNA-directed RNA polymerase I subunit RPA12, DNA-directed RNA polymerase I subunit RPA135, DNA-directed RNA polymerase I subunit RPA14, ... | | Authors: | Tafur, L, Sadian, Y, Weis, F, Muller, C.W. | | Deposit date: | 2018-09-11 | | Release date: | 2019-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The cryo-EM structure of a 12-subunit variant of RNA polymerase I reveals dissociation of the A49-A34.5 heterodimer and rearrangement of subunit A12.2.
Elife, 8, 2019
|
|
8TXA
 
 | | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Balsamo, A, Bruno, C, Edele, C, Fabian, T, Jannotta, R, Lee, R, Schryver, D, Warsaw, J, Warsaw, L, Stojanoff, V, Battaile, K, Perez, A, Bolen, R. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum
To Be Published
|
|
1FYW
 
 | | CRYSTAL STRUCTURE OF THE TIR DOMAIN OF HUMAN TLR2 | | Descriptor: | TOLL-LIKE RECEPTOR 2 | | Authors: | Xu, Y, Tao, X, Shen, B, Horng, T, Medzhitov, R, Manley, J.L, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2000-10-03 | | Release date: | 2000-11-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for signal transduction by the Toll/interleukin-1 receptor domains.
Nature, 408, 2000
|
|
6AIX
 
 | |
5OF3
 
 | |
