7MXJ
 
 | |
7MXB
 
 | |
7MXK
 
 | |
4JRG
 
 | | The 1.9A crystal structure of humanized Xenopus MDM2 with RO5313109 - a pyrrolidine MDM2 inhibitor | | Descriptor: | (3R,4R,5S)-3-(3-chlorophenyl)-4-(4-chlorophenyl)-4-cyano-N-[(3S)-3,4-dihydroxybutyl]-5-(2,2-dimethylpropyl)-D-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Graves, B.J, Janson, C.A, Lukacs, C. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of RG7388, a Potent and Selective p53-MDM2 Inhibitor in Clinical Development.
J.Med.Chem., 56, 2013
|
|
2PZ9
 
 | | Crystal structure of putative transcriptional regulator SCO4942 from Streptomyces coelicolor | | Descriptor: | Putative regulatory protein, SULFATE ION | | Authors: | Filippova, E.V, Chruszcz, M, Xu, X, Zheng, H, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-05-17 | | Release date: | 2007-06-19 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In situ proteolysis for protein crystallization and structure determination.
Nat.Methods, 4, 2007
|
|
6HHI
 
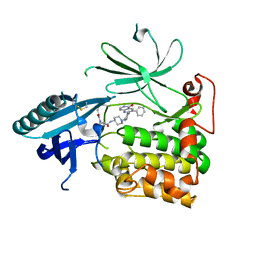 | | Crystal Structure of AKT1 in Complex with Covalent-Allosteric AKT Inhibitor 30b | | Descriptor: | RAC-alpha serine/threonine-protein kinase, ~{N}-[1-[[4-(5-oxidanylidene-3-phenyl-6~{H}-1,6-naphthyridin-2-yl)phenyl]methyl]piperidin-4-yl]-3-(propanoylamino)benzamide | | Authors: | Landel, I, Weisner, J, Mueller, M.P, Scheinpflug, R, Rauh, D. | | Deposit date: | 2018-08-28 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and chemical insights into the covalent-allosteric inhibition of the protein kinase Akt.
Chem Sci, 10, 2019
|
|
1P6L
 
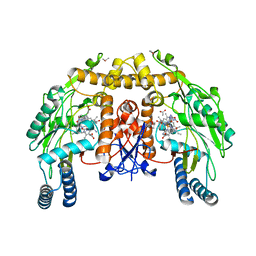 | | Bovine endothelial NOS heme domain with L-N(omega)-nitroarginine-2,4-L-diaminobutyric amide bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Flinspach, M.L, Li, H, Jamal, J, Yang, W, Huang, H, Hah, J.-M, Gomez-Vidal, J.A, Litzinger, E.A, Silverman, R.B, Poulos, T.L. | | Deposit date: | 2003-04-29 | | Release date: | 2004-01-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for dipeptide amide isoform-selective inhibition of neuronal nitric oxide synthase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
3GOY
 
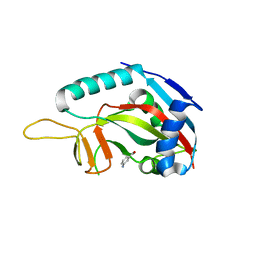 | | Crystal structure of human poly(adp-ribose) polymerase 14, catalytic fragment in complex with an inhibitor 3-aminobenzamide | | Descriptor: | 3-aminobenzamide, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Moche, M, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Schutz, P, Siponen, M.I, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors.
Nat.Biotechnol., 30, 2012
|
|
4HGJ
 
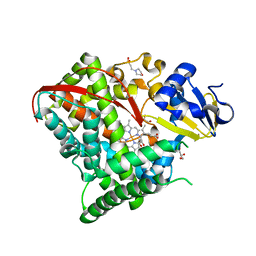 | | Crystal structure of P450 BM3 5F5 heme domain variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGG
 
 | | Crystal structure of P450 BM3 5F5R heme domain variant complexed with styrene | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGI
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset II) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
3GJJ
 
 | | crystal structure of a DNA duplex containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*(B7C)P*CP*GP*CP*G)-3' | | Authors: | Takenaka, A, Juan, E.C.M, Shimizu, S, Haraguchi, T, Xiao, M, Kurose, T, Ohkubo, A, Sekine, M, Shibata, T, Millington, C.L, Williams, D.M. | | Deposit date: | 2009-03-09 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights into the stabilizing contributions of bicyclic cytosine analogues: crystal structures of DNA duplexes containing 7,8-dihydropyridol[2,3-d]pyrimidin-2-one
To be Published
|
|
4CE8
 
 | | Perdeuterated Pseudomonas aeruginosa Lectin II complex with hydrogenated L-Fucose and Calcium | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Cuypers, M.G, Mitchell, E.P, Mossou, E, Pokorna, M, Wimmerova, M, Imberty, A, Moulin, M, Haertlein, M, Forsyth, V.T. | | Deposit date: | 2013-11-10 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Perdeuterated Pseudomonas Aeruginosa Lectin II Complex with Hydrogenated L Fucose and Calcium
To be Published
|
|
3H9U
 
 | | S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Siponen, M.I, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh Nielsen, T, Kotenyova, T, Kotzsch, A, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei
To be Published
|
|
4LYT
 
 | |
7ABS
 
 | | Structure of human DCLRE1C/Artemis in complex with DNA - re-evaluation of 6WO0 | | Descriptor: | DNA (5'-D(*CP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*GP*CP*T)-3'), Protein artemis, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-09-08 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
6UZC
 
 | |
7BT6
 
 | |
7TEO
 
 | | Cryo-EM structure of the 20S Alpha 3 Deletion proteasome core particle in complex with FUB1 | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-4, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Schnell, H.M, Hanna, J. | | Deposit date: | 2022-01-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Yeast PI31 inhibits the proteasome by a direct multisite mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TEJ
 
 | | Cryo-EM structure of the 20S Alpha 3 Deletion proteasome core particle | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-4, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Schnell, H.M, Hanna, J. | | Deposit date: | 2022-01-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Yeast PI31 inhibits the proteasome by a direct multisite mechanism.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4LGS
 
 | | Ricin A chain bound to camelid nanobody (VHH4) | | Descriptor: | Camelid nanobody (VHH4), Ricin | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M, Burshteyn, F, Cassidy, M, Gary, E, Mantis, N. | | Deposit date: | 2013-06-28 | | Release date: | 2014-06-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
J.Mol.Biol., 426, 2014
|
|
7BTB
 
 | |
4IPF
 
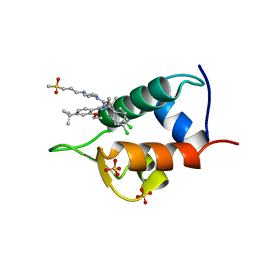 | | The 1.7A crystal structure of humanized Xenopus MDM2 with RO5045337 | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, SULFATE ION, [(4S,5R)-2-(4-tert-butyl-2-ethoxyphenyl)-4,5-bis(4-chlorophenyl)-4,5-dimethyl-4,5-dihydro-1H-imidazol-1-yl]{4-[3-(methylsulfonyl)propyl]piperazin-1-yl}methanone | | Authors: | Graves, B.J, Lukacs, C, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-01-09 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | MDM2 Small-Molecule Antagonist RG7112 Activates p53 Signaling and Regresses Human Tumors in Preclinical Cancer Models.
Cancer Res., 73, 2013
|
|
4LGP
 
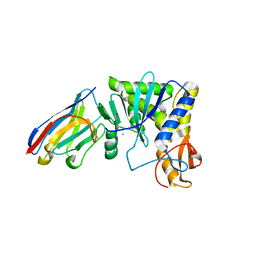 | | Ricin A chain bound to camelid nanobody (VHH1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Ricin, ... | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M, Burshteyn, F, Cassidy, M, Gary, E, Mantis, N. | | Deposit date: | 2013-06-28 | | Release date: | 2014-06-11 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
J.Mol.Biol., 426, 2014
|
|
4LHQ
 
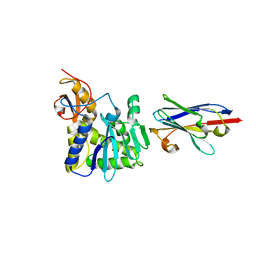 | | Ricin A chain bound to camelid nanobody (VHH8) | | Descriptor: | Camelid nanobody, Ricin | | Authors: | Rudolph, M.J, Cheung, J, Franklin, M, Burshteyn, F, Cassidy, M, Gary, E, Mantis, N. | | Deposit date: | 2013-07-01 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Ricin Toxin's Enzymatic Subunit (RTA) in Complex with Neutralizing and Non-Neutralizing Single-Chain Antibodies.
J.Mol.Biol., 426, 2014
|
|
