7E3K
 
 | | Ultrapotent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants | | Descriptor: | 13G9 heavy chain, 13G9 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Guo, H, Li, T, Liu, F, Gao, Y, Ji, X, Yang, H. | | Deposit date: | 2021-02-09 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Nat Commun, 12, 2021
|
|
8ZHQ
 
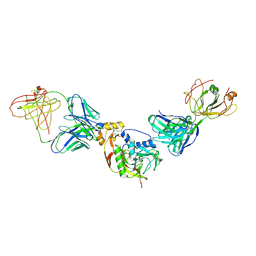 | | SFTSV Gn in complex with JK-8/12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein, ... | | Authors: | Shang, H, Guo, Y, Zhang, N, Liu, W, Li, H. | | Deposit date: | 2024-05-11 | | Release date: | 2025-01-08 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Discovery and characterization of potent broadly neutralizing antibodies from human survivors of severe fever with thrombocytopenia syndrome.
Ebiomedicine, 111, 2024
|
|
3TK9
 
 | | Crystal structure of human granzyme H | | Descriptor: | Granzyme H, SULFATE ION | | Authors: | Wang, L, Zhang, K, Wu, L, Tong, L, Sun, F, Fan, Z. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the substrate specificity of human granzyme H: the functional roles of a novel RKR motif
J.Immunol., 188, 2012
|
|
6LQO
 
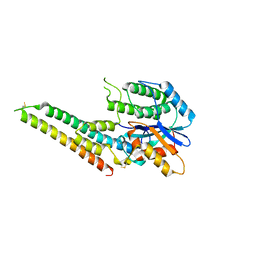 | | EBV tegument protein BBRF2/BSRF1 complex | | Descriptor: | ACETATE ION, Cytoplasmic envelopment protein 1, GLYCEROL, ... | | Authors: | He, H.P, Luo, M, Cao, Y.L, Gao, S. | | Deposit date: | 2020-01-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.0911262 Å) | | Cite: | Structure of Epstein-Barr virus tegument protein complex BBRF2-BSRF1 reveals its potential role in viral envelopment.
Nat Commun, 11, 2020
|
|
1CO0
 
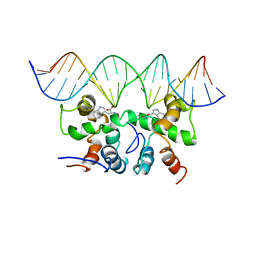 | | NMR STUDY OF TRP REPRESSOR-MTR OPERATOR DNA COMPLEX | | Descriptor: | 5'-D(*TP*GP*TP*AP*CP*CP*AP*GP*TP*AP*CP*AP*CP*GP*AP*GP*TP*AP*CP*A)-3', 5'-D(*TP*GP*TP*AP*CP*TP*CP*GP*TP*GP*TP*AP*CP*TP*GP*GP*TP*AP*CP*A)-3', TRP OPERON REPRESSOR, ... | | Authors: | Zhou, G.P, Brocchieri, L, Jardetzky, O. | | Deposit date: | 1999-05-30 | | Release date: | 2003-09-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Allostery and Induced Fit, NMR and Molecular Modeling Study of the
trp-repressor - mtr DNA complex
Structures and Mechanisms, ACS Symposium Series, 827, 2002
|
|
6INB
 
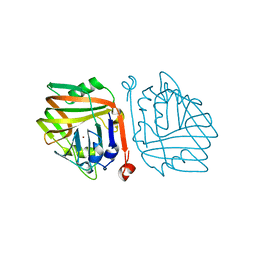 | |
5YD0
 
 | | Crystal structure of Schlafen 13 (SLFN13) N'-domain | | Descriptor: | Schlafen 8, ZINC ION | | Authors: | Yang, J.-Y, Gao, S. | | Deposit date: | 2017-09-09 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.182 Å) | | Cite: | Structure of Schlafen13 reveals a new class of tRNA/rRNA- targeting RNase engaged in translational control
Nat Commun, 9, 2018
|
|
6INC
 
 | | Crystal structure of an acetolactate decarboxylase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, Alpha-acetolactate decarboxylase, CHLORIDE ION, ... | | Authors: | Wu, W, Zhang, Q, Bartlam, M. | | Deposit date: | 2018-10-24 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural characterization of an acetolactate decarboxylase from Klebsiella pneumoniae
Biochem. Biophys. Res. Commun., 509, 2019
|
|
7FH9
 
 | |
1EBY
 
 | | HIV-1 protease in complex with the inhibitor BEA369 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DIBENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
7FH4
 
 | | Chlorovirus PBCV-1 bi-functional dCMP/dCTP deaminase bi-DCD | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CMP/dCMP-type deaminase domain-containing protein, ... | | Authors: | She, Z. | | Deposit date: | 2021-07-29 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structural basis of a multi-functional deaminase in chlorovirus PBCV-1.
Arch.Biochem.Biophys., 727, 2022
|
|
8IMS
 
 | | Crystal structure of TRAF7 coiled-coil domain | | Descriptor: | E3 ubiquitin-protein ligase TRAF7 | | Authors: | Hu, R, Lin, L, Lu, Q. | | Deposit date: | 2023-03-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of TRAF7 coiled-coil trimer provides insight into its function in zebrafish embryonic development.
J Mol Cell Biol, 16, 2024
|
|
8IJC
 
 | | NMR solution structure of the 1:1 complex of a platinum(II) ligand L1-transpt covalently bound to a G-quadruplex MYT1L | | Descriptor: | G-quadruplex DNA MYT1L, Pt(NH3)2(2-(pyridin-4-ylmethyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) | | Authors: | Liu, L.-Y, Mao, Z.-W. | | Deposit date: | 2023-02-27 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Organic-Platinum Hybrids for Covalent Binding of G-Quadruplexes: Structural Basis and Application to Cancer Immunotherapy.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6AAU
 
 | | Solution Structure for m62A helix 45 in 3' end of 12S rRNA | | Descriptor: | RNA (24-mer) | | Authors: | Liu, X, Wu, P. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2025-02-12 | | Method: | SOLUTION NMR | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
8TU6
 
 | | CryoEM structure of PI3Kalpha | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Valverde, R, Shi, H, Holliday, M. | | Deposit date: | 2023-08-15 | | Release date: | 2023-11-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Discovery and Clinical Proof-of-Concept of RLY-2608, a First-in-Class Mutant-Selective Allosteric PI3K alpha Inhibitor That Decouples Antitumor Activity from Hyperinsulinemia.
Cancer Discov, 14, 2024
|
|
4YO4
 
 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment phthalazine | | Descriptor: | ACETATE ION, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Roy, S.M, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
4YPD
 
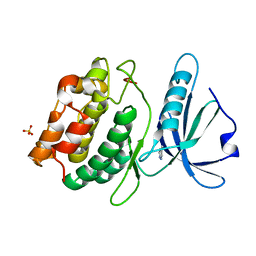 | | Crystal Structure of DAPK1 catalytic domain in complex with the hinge binding fragment 4-methylpyridazine | | Descriptor: | 4-methylpyridazine, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Roy, S.M, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of DAPK1 catalytic domain in complex with hinge binding fragments
To Be Published
|
|
6OLU
 
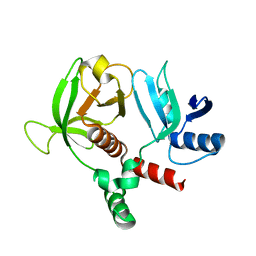 | | RIAM RA-PH core structure in the P212121 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-04-17 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation of RIAM by src promotes integrin activation by unmasking the PH domain of RIAM.
Structure, 29, 2021
|
|
6O6H
 
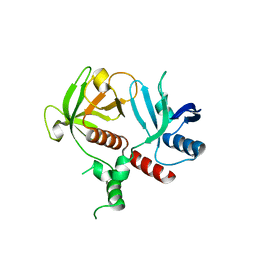 | | RIAM cc-RA-PH structure in the P21212 space group | | Descriptor: | Amyloid beta A4 precursor protein-binding family B member 1-interacting protein | | Authors: | Wu, J. | | Deposit date: | 2019-03-06 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphorylation of RIAM by Src Promotes Integrin Activation by Unmasking the PH Domain of RIAM.
Structure, 2020
|
|
6B88
 
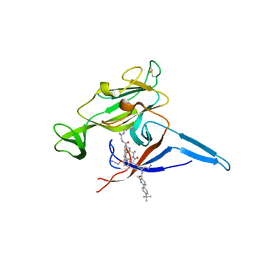 | | E. coli LepB in complex with GNE0775 ((4S,7S,10S)-10-((S)-4-amino-2-(2-(4-(tert-butyl)phenyl)-4-methylpyrimidine-5-carboxamido)-N-methylbutanamido)-16,26-bis(2-aminoethoxy)-N-(2-iminoethyl)-7-methyl-6,9-dioxo-5,8-diaza-1,2(1,3)-dibenzenacyclodecaphane-4-carboxamide) | | Descriptor: | (8S,11S,14S)-14-{[(2S)-4-amino-2-{[2-(4-tert-butylphenyl)-4-methylpyrimidine-5-carbonyl]amino}butanoyl](methyl)amino}-3,18-bis(2-aminoethoxy)-N-[(2Z)-2-iminoethyl]-11-methyl-10,13-dioxo-9,12-diazatricyclo[13.3.1.1~2,6~]icosa-1(19),2(20),3,5,15,17-hexaene-8-carboxamide, PENTAETHYLENE GLYCOL, Signal peptidase I | | Authors: | Murray, J.M, Rouge, L. | | Deposit date: | 2017-10-05 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Optimized arylomycins are a new class of Gram-negative antibiotics.
Nature, 561, 2018
|
|
7XCZ
 
 | |
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | Authors: | Liu, Z, Lui, S, Gao, Y. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | Authors: | Liu, Z, Liu, S, Gao, Y.Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDA
 
 | |
7XDL
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7208 and BA7125 fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7125 fab, ... | | Authors: | Liu, Z, Liu, S, Yuanzhu, G. | | Deposit date: | 2022-03-27 | | Release date: | 2023-03-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
