7YSL
 
 | | Crystal structure of D-Cysteine desulfhydrase with a trapped PLP-pyruvate geminal diamine | | Descriptor: | 1,2-ETHANEDIOL, D-Cysteine desulfhydrase, FORMIC ACID | | Authors: | Zhang, X, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-08-12 | | Release date: | 2022-12-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Characterization and structural basis of D-cysteine desulfhydrase from Pectobacterium atrosepticum
Tetrahedron, 2022
|
|
8WQ2
 
 | |
8WQ4
 
 | |
8WKP
 
 | |
6L6X
 
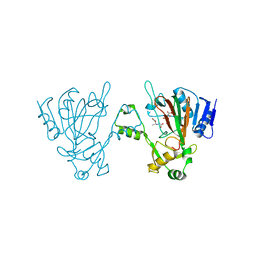 | | The structure of ScoE with substrate | | Descriptor: | (3~{R})-3-(2-hydroxy-2-oxoethylamino)butanoic acid, D(-)-TARTARIC ACID, FE (II) ION, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-10-29 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6L86
 
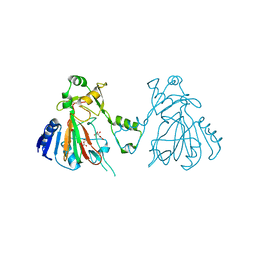 | | The structure of SfaA | | Descriptor: | (2S)-2-hydroxybutanedioic acid, D-MALATE, FE (II) ION, ... | | Authors: | Chen, T.Y, Chen, J, Zhou, J, Chang, W. | | Deposit date: | 2019-11-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Pathway from N-Alkylglycine to Alkylisonitrile Catalyzed by Iron(II) and 2-Oxoglutarate-Dependent Oxygenases.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8HKO
 
 | |
8HKS
 
 | | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib(BGB-290) | | Descriptor: | (2R)-14-fluoro-2-methyl-6,9,10,19-tetrazapentacyclo[14.2.1.02,6.08,18.012,17]nonadeca-1(18),8,12(17),13,15-pentaen-11-one, GLYCEROL, Poly [ADP-ribose] polymerase 2 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2022-11-28 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mutated human ADP-ribosyltransferase 2 (PARP2) catalytic domain bound to Pamiparib
To Be Published
|
|
8HLJ
 
 | |
8HLR
 
 | |
8HLQ
 
 | |
8HKN
 
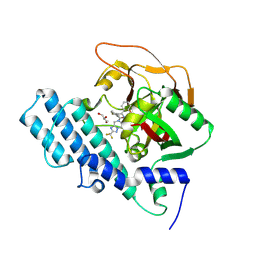 | |
2RKK
 
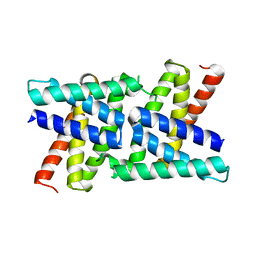 | |
2RKL
 
 | | Crystal Structure of S.cerevisiae Vta1 C-terminal domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Vacuolar protein sorting-associated protein VTA1 | | Authors: | Xiao, J, Xia, H, Zhou, J, Xu, Z. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of vta1 function in the multivesicular body sorting pathway.
Dev.Cell, 14, 2008
|
|
4RNC
 
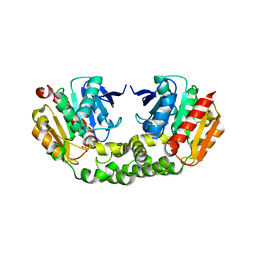 | | Crystal structure of an esterase RhEst1 from Rhodococcus sp. ECU1013 | | Descriptor: | Esterase, PHOSPHATE ION | | Authors: | Dou, S, Kong, X.D, Xu, J.H, Zhou, J. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Substrate channel evolution of an esterase for the synthesis of Cilastatin
CATALYSIS SCIENCE AND TECHNOLOGY, 5, 2015
|
|
6L29
 
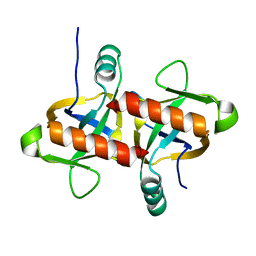 | | The structure of the MazF-mt1 mutant | | Descriptor: | mRNA interferase | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-10-02 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3000052 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6KYT
 
 | | The structure of the M. tb toxin MazEF-mt1 complex | | Descriptor: | Antitoxin MazE9, Endoribonuclease MazF9 | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-09-20 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.00101161 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6KYS
 
 | | The structure of the M. tb toxin MazF-mt1 | | Descriptor: | Endoribonuclease MazF9 | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-09-20 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.200414 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6L2A
 
 | | A mutant form of M. tb toxin MazEF-mt1 | | Descriptor: | mRNA interferase | | Authors: | Xie, W, Chen, R, Zhou, J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.90044665 Å) | | Cite: | Conserved Conformational Changes in the Regulation ofMycobacterium tuberculosisMazEF-mt1.
Acs Infect Dis., 6, 2020
|
|
6LM2
 
 | |
6LPS
 
 | |
1YIS
 
 | | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase | | Descriptor: | SULFATE ION, adenylosuccinate lyase | | Authors: | Symersky, J, Schormann, N, Lu, S, Zhang, Y, Karpova, E, Qiu, S, Huang, W, Cao, Z, Zhou, J, Luo, M, Arabshahi, A, McKinstry, A, Luan, C.-H, Luo, D, Johnson, D, An, J, Tsao, J, Delucas, L, Shang, Q, Gray, R, Li, S, Bray, T, Chen, Y.-J, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-01-12 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural genomics of Caenorhabditis elegans: adenylosuccinate lyase
To be Published
|
|
3WFV
 
 | | HIV-1 CRF07 gp41 | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Du, J, Xue, H, Ma, J, Liu, F, Zhou, J, Shao, Y, Qiao, W, Liu, X. | | Deposit date: | 2013-07-24 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of HIV CRF07 B'/C gp41 reveals a hyper-mutant site in the middle of HR2 heptad repeat
Virology, 446, 2013
|
|
4JFG
 
 | | Crystal structure of sfGFP-66-HqAla | | Descriptor: | CESIUM ION, Green fluorescent protein, quinolin-8-ol | | Authors: | Wang, J, Liu, X, Li, J, Zhang, W, Hu, M, Zhou, J. | | Deposit date: | 2013-02-28 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Significant expansion of the fluorescent protein chromophore through the genetic incorporation of a metal-chelating unnatural amino acid.
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
1XKQ
 
 | | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, short-chain reductase family member (5D234) | | Authors: | Schormann, N, Zhou, J, Karpova, E, Zhang, Y, Symersky, J, Bunzel, B, Huang, W.-Y, Arabshahi, A, Qiu, S, Luan, C.-H, Gray, R, Carson, M, Tsao, J, Luo, M, Johnson, D, Lu, S, Lin, G, Luo, D, Cao, Z, Li, S, McKinstry, A, Shang, Q, Chen, Y.-J, Bray, T, Nagy, L, DeLucas, L, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-29 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Short-Chain Dehydrogenase/Reductase of unknown Function from Caenorhabditis Elegans with Cofactor
To be Published
|
|
