3N27
 
 | |
3EEM
 
 | | Candida glabrata Dihydrofolate Reductase complexed with 2,4-diamino-5-[3-methyl-3-(3-methoxy-5-(2,6-dimethylphenyl)phenyl)prop-1-ynyl]-6-methylpyrimidine(UCP111D26M) and NADPH | | Descriptor: | 5-[(3R)-3-(5-methoxy-2',6'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Probing the active site of Candida glabrata dihydrofolate reductase with high resolution crystal structures and the synthesis of new inhibitors
Chem.Biol.Drug Des., 73, 2009
|
|
3EEK
 
 | |
3EEL
 
 | | Candida glabrata Dihydrofolate Reductase complexed with 2,4-diamino-5-[3-methyl-3-(3-methoxy-5-(3,5-dimethylphenyl)phenyl)prop-1-ynyl]-6-methylpyrimidine(UCP11153TM) and NADPH | | Descriptor: | 5-[(3R)-3-(5-methoxy-3',5'-dimethylbiphenyl-3-yl)but-1-yn-1-yl]-6-methylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, J, Anderson, A. | | Deposit date: | 2008-09-04 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the active site of Candida glabrata dihydrofolate reductase with high resolution crystal structures and the synthesis of new inhibitors
Chem.Biol.Drug Des., 73, 2009
|
|
3EEJ
 
 | |
1SUW
 
 | | Crystal structure of a NAD kinase from Archaeoglobus fulgidus in complex with its substrate and product: Insights into the catalysis of NAD kinase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable inorganic polyphosphate/ATP-NAD kinase | | Authors: | Liu, J, Lou, Y, Yokota, H, Adams, P.D, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of an NAD Kinase from Archaeoglobus fulgidus in Complex with ATP, NAD, or NADP
J.Mol.Biol., 354, 2005
|
|
4F7T
 
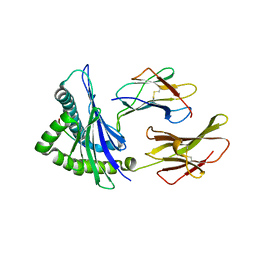 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PB1 (498-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
4F7M
 
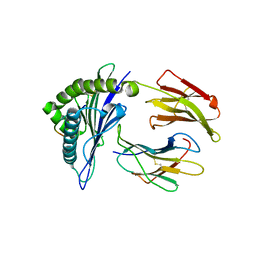 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PA (649-658) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, George, F.G. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
4F7P
 
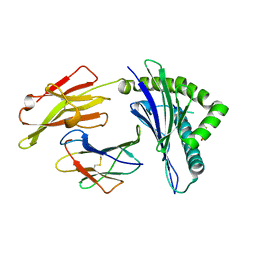 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009H1N1 PB1 (496-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
3OMI
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OMA
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3TO2
 
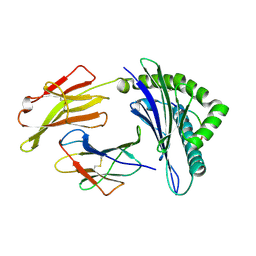 | | Structure of HLA-A*0201 complexed with peptide Md3-C9 derived from a clustering region of restricted cytotoxic T lymphocyte epitope from SARS-CoV M protein | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Md3-C9 peptide derived from Membrane glycoprotein | | Authors: | Liu, J, Qi, J, Gao, F, Yan, J, Gao, G.F. | | Deposit date: | 2011-09-03 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional and Structural Definition of a Clustering Region of HLA-A2-restricted Cytotoxic T Lymphocyte Epitopes
Sci.Technology Rev., 29, 2011
|
|
3OM3
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with K362M mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-26 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3OXS
 
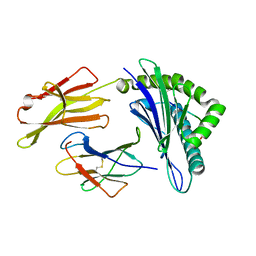 | | Crystal Structure of HLA A*02:07 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-22 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
3OXR
 
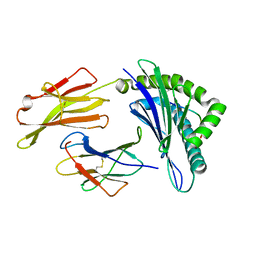 | | Crystal Structure of HLA A*02:06 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
3OX8
 
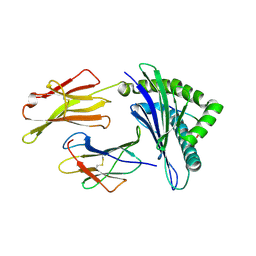 | | Crystal Structure of HLA A*02:03 Bound to HBV Core 18-27 | | Descriptor: | 10mer peptide from Pre-core-protein, Beta-2-microglobulin, MHC class I antigen | | Authors: | Liu, J, Chen, Y, Lai, L, Ren, E. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural insights into the binding of hepatitis B virus core peptide to HLA-A2 alleles: Towards designing better vaccines.
Eur.J.Immunol., 41, 2011
|
|
1AIL
 
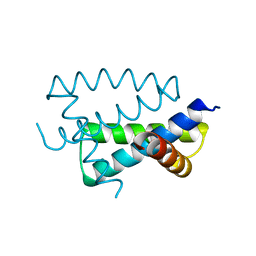 | | N-TERMINAL FRAGMENT OF NS1 PROTEIN FROM INFLUENZA A VIRUS | | Descriptor: | NONSTRUCTURAL PROTEIN NS1 | | Authors: | Liu, J, Lynch, P.A, Chien, C, Montelione, G.T, Krug, R.M, Berman, H.M. | | Deposit date: | 1997-04-21 | | Release date: | 1997-10-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein.
Nat.Struct.Biol., 4, 1997
|
|
3OMN
 
 | | Catalytic core subunits (I and II) of cytochrome C oxidase from Rhodobacter sphaeroides with D132A mutation in the reduced state | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, CADMIUM ION, CALCIUM ION, ... | | Authors: | Liu, J, Qin, L, Ferguson-Miller, S. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2HY6
 
 | | A seven-helix coiled coil | | Descriptor: | General control protein GCN4, HEXANE-1,6-DIOL | | Authors: | Liu, J, Zheng, Q, Deng, Y, Cheng, C.S, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-08-04 | | Release date: | 2006-10-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A seven-helix coiled coil.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GUV
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
2GUS
 
 | | Conformational Transition between Four- and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction | | Descriptor: | Major outer membrane lipoprotein | | Authors: | Liu, J, Zheng, Q, Deng, Y, Kallenbach, N.R, Lu, M. | | Deposit date: | 2006-05-01 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Conformational Transition between Four and Five-stranded Phenylalanine Zippers Determined by a Local Packing Interaction.
J.Mol.Biol., 361, 2006
|
|
2IPZ
 
 | | A Parallel Coiled-Coil Tetramer with Offset Helices | | Descriptor: | GLYCEROL, General control protein GCN4, ISOPROPYL ALCOHOL | | Authors: | Liu, J, Lu, M. | | Deposit date: | 2006-10-12 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A Parallel Coiled-Coil Tetramer with Offset Helices.
Biochemistry, 45, 2006
|
|
2MTP
 
 | |
3TCP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC569 | | Descriptor: | 1-[(trans-4-aminocyclohexyl)methyl]-N-butyl-3-(4-fluorophenyl)-1H-pyrazolo[3,4-d]pyrimidin-6-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Liu, J, Yang, C, Simpson, C, DeRyckere, D, Van Deusen, A, Miley, M, Kireev, D.B, Norris-Drouin, J, Sather, S, Hunter, D, Patel, H.S, Janzen, W.P, Machius, M, Johnson, G, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2011-08-09 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of Novel Small Molecule Mer Kinase Inhibitors for the Treatment of Pediatric Acute Lymphoblastic Leukemia.
ACS Med Chem Lett, 3, 2012
|
|
2LKO
 
 | | Structural Basis of Phosphoinositide Binding to Kindlin-2 Pleckstrin Homology Domain in Regulating Integrin Activation | | Descriptor: | Fermitin family homolog 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Liu, J, Fukuda, K, Xu, Z. | | Deposit date: | 2011-10-17 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of phosphoinositide binding to kindlin-2 protein pleckstrin homology domain in regulating integrin activation.
J.Biol.Chem., 286, 2011
|
|
