5B3S
 
 | | Bovine heart cytochrome c oxidase in the carbon monoxide-bound mixed-valence state at 1.68 angstrom resolution (50 K) | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Shinzawa-Ito, K, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2016-03-11 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Bovine heart cytochrome c oxidase in the carbon monoxide-bound mixed-valence state at 1.68 angstrom resolution (50 K)
To Be Published
|
|
5X1B
 
 | | CO bound cytochrome c oxidase at 20 nsec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2017-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
5X19
 
 | | CO bound cytochrome c oxidase at 100 micro sec after pump laser irradiation to release CO from O2 reduction center | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Kubo, M, Baba, S, Yamashita, K, Hirata, K, Ueno, G, Nomura, T, Kimura, T, Shinzawa-Itoh, K, Baba, J, Hatano, K, Eto, Y, Miyamoto, A, Murakami, H, Kumasaka, T, Owada, S, Tono, K, Yabashi, M, Yamaguchi, Y, Yanagisawa, S, Sakaguchi, M, Ogura, T, Komiya, R, Yan, J, Yamashita, E, Yamamoto, M, Ago, H, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A nanosecond time-resolved XFEL analysis of structural changes associated with CO release from cytochrome c oxidase.
Sci Adv, 3, 2017
|
|
2HI7
 
 | | Crystal structure of DsbA-DsbB-ubiquinone complex | | Descriptor: | Disulfide bond formation protein B, Thiol:disulfide interchange protein dsbA, UBIQUINONE-1, ... | | Authors: | Inaba, K, Murakami, S, Suzuki, M, Nakagawa, A, Yamashita, E, Okada, K, Ito, K. | | Deposit date: | 2006-06-29 | | Release date: | 2006-12-05 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the DsbB-DsbA Complex Reveals a Mechanism of Disulfide Bond Generation
Cell(Cambridge,Mass.), 127, 2006
|
|
3J9W
 
 | | Cryo-EM structure of the Bacillus subtilis MifM-stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein bS16, ... | | Authors: | Sohmen, D, Chiba, S, Shimokawa-Chiba, N, Innis, C.A, Berninghausen, O, Beckmann, R, Ito, K, Wilson, D.N. | | Deposit date: | 2015-03-16 | | Release date: | 2015-04-29 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling.
Nat Commun, 6, 2015
|
|
1IRE
 
 | | Crystal Structure of Co-type nitrile hydratase from Pseudonocardia thermophila | | Descriptor: | COBALT (II) ION, Nitrile Hydratase | | Authors: | Miyanaga, A, Fushinobu, S, Ito, K, Wakagi, T. | | Deposit date: | 2001-10-01 | | Release date: | 2002-10-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cobalt-containing nitrile hydratase.
Biochem.Biophys.Res.Commun., 288, 2001
|
|
4BEH
 
 | | Solution structure of human ribosomal protein P1.P2 heterodimer | | Descriptor: | 60S ACIDIC RIBOSOMAL PROTEIN P1, 60S ACIDIC RIBOSOMAL PROTEIN P2 | | Authors: | Lee, K.M, Yusa, K, Chu, L.O, Wing-Heng Yu, C, Shaw, P.C, Oono, M, Miyoshi, T, Ito, K, Wong, K.B, Uchiumi, T. | | Deposit date: | 2013-03-10 | | Release date: | 2013-08-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human P1P2 Heterodimer Provides Insights Into the Role of Eukaryotic Stalk in Recruiting the Ribosome-Inactivating Protein Trichosanthin to the Ribosome.
Nucleic Acids Res., 41, 2013
|
|
2IPC
 
 | | Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer | | Descriptor: | Preprotein translocase SecA subunit | | Authors: | Vassylyev, D.G, Mori, H, Vassylyeva, M.N, Tsukazaki, T, Kimura, Y, Tahirov, T.H, Ito, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Translocation ATPase SecA from Thermus thermophilus Reveals a Parallel, Head-to-Head Dimer.
J.Mol.Biol., 364, 2006
|
|
5AWH
 
 | | Rhodobacter sphaeroides Argonaute in complex with guide RNA/target DNA heteroduplex | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*A)-3'), MAGNESIUM ION, RNA (5'-D(P*UP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3'), ... | | Authors: | Miyoshi, T, Ito, K, Murakami, R, Uchiumi, T. | | Deposit date: | 2015-07-03 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute.
Nat Commun, 7, 2016
|
|
5YT0
 
 | | Crystal structure of the complex of archaeal ribosomal stalk protein aP1 and archaeal translation initiation factor aIF5B | | Descriptor: | Archaeal ribosomal stalk protein aP1, GUANOSINE-5'-DIPHOSPHATE, Probable translation initiation factor IF-2 | | Authors: | Murakami, R, Singh, C.R, Morris, J, Tang, L, Harmon, I, Miyoshi, T, Ito, K, Asano, K, Uchiumi, T. | | Deposit date: | 2017-11-16 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Interaction between the Ribosomal Stalk Proteins and Translation Initiation Factor 5B Promotes Translation Initiation
Mol. Cell. Biol., 38, 2018
|
|
5FG3
 
 | | Crystal structure of GDP-bound aIF5B from Aeropyrum pernix | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Probable translation initiation factor IF-2 | | Authors: | Murakami, R, Miyoshi, T, Uchiumi, T, Ito, K. | | Deposit date: | 2015-12-20 | | Release date: | 2016-05-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of translation initiation factor 5B from the crenarchaeon Aeropyrum pernix.
Proteins, 84, 2016
|
|
1J2T
 
 | | Creatininase Mn | | Descriptor: | MANGANESE (II) ION, SULFATE ION, ZINC ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1J2U
 
 | | Creatininase Zn | | Descriptor: | SULFATE ION, ZINC ION, creatinine amidohydrolase | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-01-11 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
5YV5
 
 | | Crystal structure of the complex of archaeal ribosomal stalk protein aP1 and archaeal ribosome recycling factor aABCE1. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATPase RIL, Archaeal ribosomal stalk protein aP1, ... | | Authors: | Imai, H, Abe, T, Miyoshi, T, Nishikawa, S, Ito, K, Uchiumi, T. | | Deposit date: | 2017-11-24 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The ribosomal stalk protein is crucial for the action of the conserved ATPase ABCE1
Nucleic Acids Res., 46, 2018
|
|
1KOL
 
 | | Crystal structure of formaldehyde dehydrogenase | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ZINC ION, ... | | Authors: | Tanaka, N, Kusakabe, Y, Ito, K, Yoshimoto, T, Nakamura, K.T. | | Deposit date: | 2001-12-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Formaldehyde Dehydrogenase from Pseudomonas putida: the Structural Origin of the Tightly Bound Cofactor in Nicotinoprotein Dehydrogenases
J.mol.biol., 324, 2002
|
|
5ZX8
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Thermus thermophilus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CITRATE ANION, Peptidyl-tRNA hydrolase | | Authors: | Matsumoto, A, Uehara, U, Shimizu, Y, Ueda, T, Uchiumi, T, Ito, K. | | Deposit date: | 2018-05-18 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure of peptidyl-tRNA hydrolase from Thermus thermophilus.
Proteins, 87, 2019
|
|
6L37
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L36
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6L38
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW9662-gemfibrozil co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-chloro-5-nitro-N-phenylbenzamide, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-10-09 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.761 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX4
 
 | | X-ray structure of human PPARalpha ligand binding domain-fenofibric acid co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-[4-(4-chlorobenzene-1-carbonyl)phenoxy]-2-methylpropanoic acid, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX9
 
 | | X-ray structure of human PPARalpha ligand binding domain-arachidonic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | ARACHIDONIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX5
 
 | | X-ray structure of human PPARalpha ligand binding domain-ciprofibrate co-crystals obtained by delipidation and co-crystallization | | Descriptor: | 2-{4-[(1S)-2,2-dichlorocyclopropyl]phenoxy}-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX6
 
 | | X-ray structure of human PPARalpha ligand binding domain-palmitic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, PALMITIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LXA
 
 | | X-ray structure of human PPARalpha ligand binding domain-eicosapentaenoic acid (EPA) co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
6LX8
 
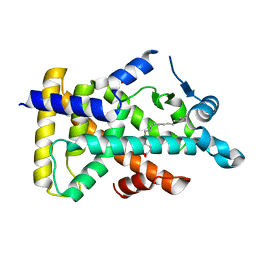 | | X-ray structure of human PPARalpha ligand binding domain-oleic acid co-crystals obtained by delipidation and cross-seeding | | Descriptor: | GLYCEROL, OLEIC ACID, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
