8SG1
 
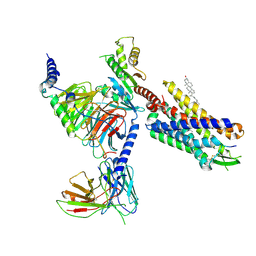 | | Cryo-EM structure of CMKLR1 signaling complex | | Descriptor: | CHOLESTEROL, Chemerin 9, Chemerin-like receptor 1, ... | | Authors: | Zhang, X, Zhang, C. | | Deposit date: | 2023-04-11 | | Release date: | 2023-11-01 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis of G protein-Coupled receptor CMKLR1 activation and signaling induced by a chemerin-derived agonist.
Plos Biol., 21, 2023
|
|
6AH0
 
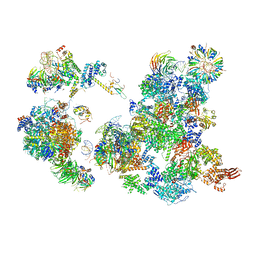 | | The Cryo-EM Structure of the Precusor of Human Pre-catalytic Spliceosome (pre-B complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-15 | | Release date: | 2018-11-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
6AHD
 
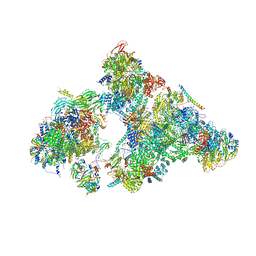 | | The Cryo-EM Structure of Human Pre-catalytic Spliceosome (B complex) at 3.8 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Brr2, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Zhan, X, Yan, C, Zhang, X, Shi, Y. | | Deposit date: | 2018-08-17 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the human pre-catalytic spliceosome and its precursor spliceosome.
Cell Res., 28, 2018
|
|
5Z58
 
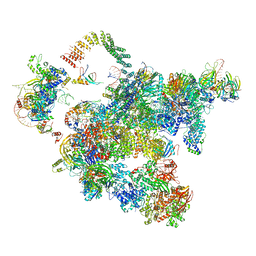 | | Cryo-EM structure of a human activated spliceosome (early Bact) at 4.9 angstrom. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Yan, C, Zhan, X, Li, L, Lei, J, Shi, Y. | | Deposit date: | 2018-01-17 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of the human activated spliceosome in three conformational states.
Cell Res., 28, 2018
|
|
6LSV
 
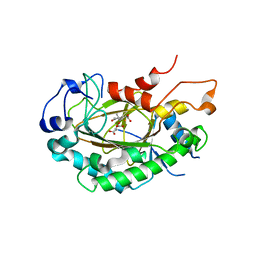 | | Crystal structure of JOX2 in complex with 2OG, Fe, and JA | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Probable 2-oxoglutarate-dependent dioxygenase At5g05600, ... | | Authors: | Zhang, X, Wang, D, Liu, J. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structure-guided analysis of Arabidopsis JASMONATE-INDUCED OXYGENASE (JOX) 2 reveals key residues for recognition of jasmonic acid substrate by plant JOXs.
Mol Plant, 14, 2021
|
|
1H0C
 
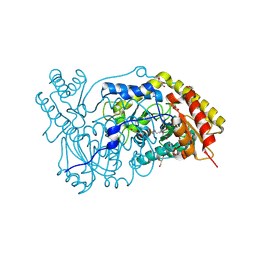 | | The crystal structure of human alanine:glyoxylate aminotransferase | | Descriptor: | (AMINOOXY)ACETIC ACID, GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Zhang, X, Danpure, C.J, Roe, S.M, Pearl, L.H. | | Deposit date: | 2002-06-17 | | Release date: | 2003-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Alanine:Glyoxylate Aminotransferase and the Relationship between Genotype and Enzymatic Phenotype in Primary Hyperoxaluria Type 1.
J.Mol.Biol., 331, 2003
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
9J0X
 
 | |
9J10
 
 | |
9J0Y
 
 | |
9J0Z
 
 | |
9ITO
 
 | | Chloroflexus aurantiacus ATP synthase, state 2, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITW
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 1, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITN
 
 | | Chloroflexus aurantiacus ATP synthase, state 1, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITX
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 2, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITU
 
 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITT
 
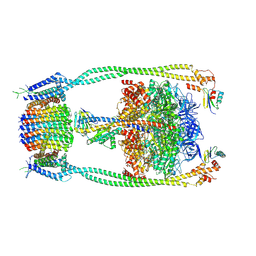 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITS
 
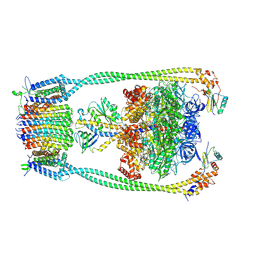 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITK
 
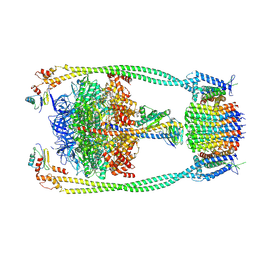 | | Chloroflexus aurantiacus ATP synthase, state 2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITR
 
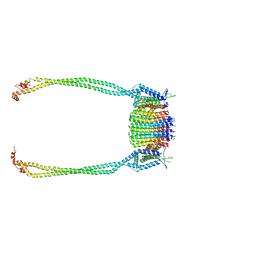 | | Chloroflexus aurantiacus ATP synthase, state 3, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IU0
 
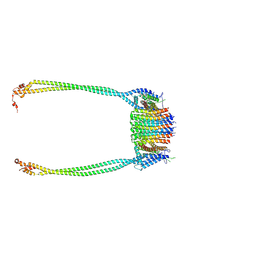 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3, focused refinement of FO and peripheral stalk | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (5.18 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITL
 
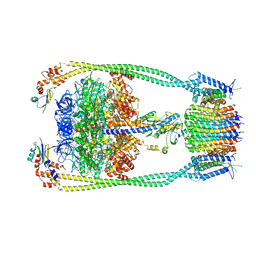 | | Chloroflexus aurantiacus ATP synthase, state 3 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITQ
 
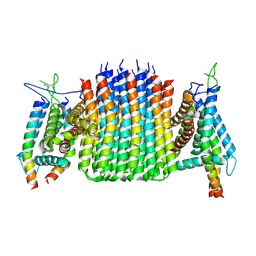 | | Chloroflexus aurantiacus ATP synthase, state 3, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITZ
 
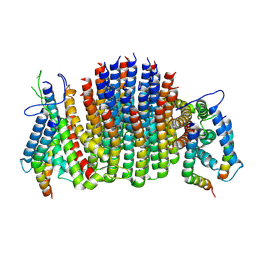 | | Chloroflexus aurantiacus ADP-bound ATP synthase, state 3, focused refinement of FO | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITJ
 
 | | Chloroflexus aurantiacus ATP synthase, state 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ATP synthase gamma chain, ... | | Authors: | Zhang, X, Wu, J, Xu, X. | | Deposit date: | 2024-07-20 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structure of ATP synthase from an early photosynthetic bacterium Chloroflexus aurantiacus.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
