5ZGM
 
 | | Crystal Structure of Parvalbumin SPVI, the Major Allergens in Mustelus griseus | | Descriptor: | CALCIUM ION, Parvalbumin SPVI, SULFATE ION | | Authors: | Yang, R.Q, Chen, Y.L, Jin, T.C, Cao, M.J. | | Deposit date: | 2018-03-09 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
J. Agric. Food Chem., 66, 2018
|
|
5ZH6
 
 | | Crystal structure of Parvalbumin SPV-II of Mustelus griseus | | Descriptor: | CALCIUM ION, Parvalbumin SPV-II, SULFATE ION | | Authors: | Yang, R.Q, Chen, Y.L, Jin, T.C, Cao, M.J. | | Deposit date: | 2018-03-11 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Purification, Characterization, and Crystal Structure of Parvalbumins, the Major Allergens in Mustelus griseus.
J. Agric. Food Chem., 66, 2018
|
|
7DV4
 
 | | Crystal structure of anti-CTLA-4 VH domain in complex with human CTLA-4 | | Descriptor: | 1,2-ETHANEDIOL, 4003-1(VH), Cytotoxic T-lymphocyte protein 4 | | Authors: | Li, H, Gan, X, He, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | An anti-CTLA-4 heavy chain-only antibody with enhanced T reg depletion shows excellent preclinical efficacy and safety profile.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3B5I
 
 | | Crystal structure of Indole-3-acetic Acid Methyltransferase | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase-like protein | | Authors: | Ferrer, J.-L, Noel, J.P. | | Deposit date: | 2007-10-26 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural, Biochemical, and Phylogenetic Analyses Suggest That Indole-3-Acetic Acid Methyltransferase Is an Evolutionarily Ancient Member of the SABATH Family.
Plant Physiol., 146, 2008
|
|
8YEO
 
 | | Type I-FHNH Cascade-dsDNA R-loop complex | | Descriptor: | 60-nt crRNA, Cas5f, Cas6f, ... | | Authors: | Li, Z. | | Deposit date: | 2024-02-22 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanisms for HNH-mediated target DNA cleavage in type I CRISPR-Cas systems.
Mol.Cell, 84, 2024
|
|
6LQL
 
 | | Complex structure of CHAO with product from Erythrobacteraceae bacterium | | Descriptor: | 1-[(4-methoxyphenyl)methyl]-3,4,5,6,7,8-hexahydroisoquinoline, Amine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Huang, Z.D. | | Deposit date: | 2020-01-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Asymmetric Synthesis of a Key Dextromethorphan Intermediate and Its Analogues Enabled by a New Cyclohexylamine Oxidase: Enzyme Discovery, Reaction Development, and Mechanistic Insight.
J.Org.Chem., 85, 2020
|
|
6LQC
 
 | | Crystal structure of Cyclohexylamine Oxidase from Erythrobacteraceae bacterium | | Descriptor: | Cyclohexylamine Oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Huang, Z.D. | | Deposit date: | 2020-01-13 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Asymmetric Synthesis of a Key Dextromethorphan Intermediate and Its Analogues Enabled by a New Cyclohexylamine Oxidase: Enzyme Discovery, Reaction Development, and Mechanistic Insight.
J.Org.Chem., 85, 2020
|
|
7DML
 
 | |
7E9A
 
 | | Crystal structure of KPC-2 in complex with (S)-2-(1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-3-yl)acrylic acid (4a-(S)) | | Descriptor: | 2-[(3S)-1-oxidanyl-3H-2,1-benzoxaborol-3-yl]prop-2-enoic acid, ACETIC ACID, Beta-lactamase, ... | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2021-03-03 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and enantioselective synthesis of 3-( alpha-acrylic acid) benzoxaboroles to combat carbapenemase resistance.
Chem.Commun.(Camb.), 57, 2021
|
|
5IT3
 
 | | Swirm domain of human Lsd1 | | Descriptor: | Lysine-specific histone demethylase 1A, MAGNESIUM ION | | Authors: | Jeffrey, P.D, Yuan, P. | | Deposit date: | 2016-03-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tlx-interacting peptide of Lsd1 inhibits the proliferation of brain tumor stem cells
To Be Published
|
|
7JTG
 
 | |
7JTF
 
 | |
6J72
 
 | | Crystal structure of IniA from Mycobacterium smegmatis with GTP bound | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Isoniazid inducible gene protein IniA, L(+)-TARTARIC ACID, ... | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
6J73
 
 | | Crystal structure of IniA from Mycobacterium smegmatis | | Descriptor: | Isoniazid inducible gene protein IniA | | Authors: | Wang, M.F, Guo, X.Y, Hu, J.J, Li, J, Rao, Z.H. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.211 Å) | | Cite: | Mycobacterial dynamin-like protein IniA mediates membrane fission.
Nat Commun, 10, 2019
|
|
5YZP
 
 | | Crystal structure of p204 HINa domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Ifi204 | | Authors: | Jin, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.581 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
5YZW
 
 | | Crystal structure of p204 HINb domain | | Descriptor: | Ifi204 | | Authors: | Jin, T. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
5ZTC
 
 | |
5Z7D
 
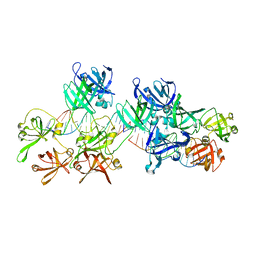 | | p204HINab-dsDNA complex structure | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*CP*AP*GP*AP*AP*AP*GP*AP*GP*AP*GP*C)-3'), Interferon-activable protein 204 | | Authors: | Jin, T, Jiang, J, Xiao, T.S. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
8I6K
 
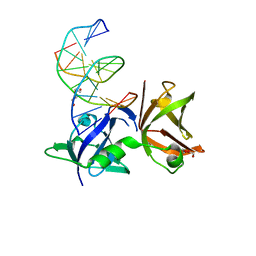 | | Structure of hMNDA HIN with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-01-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of dsDNA recognition by the hMNDA HIN domain: New insights into the DNA-binding model of a PYHIN protein.
Int.J.Biol.Macromol., 245, 2023
|
|
7ESV
 
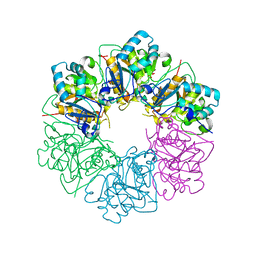 | |
7ESQ
 
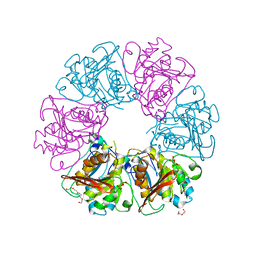 | |
7ESP
 
 | |
7ESO
 
 | |
3UO9
 
 | | Crystal Structure of Human GAC in Complex with Glutamate and BPTES | | Descriptor: | GLYCEROL, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wei, W, Hurov, J.B. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
3UNW
 
 | | Crystal Structure of Human GAC in Complex with Glutamate | | Descriptor: | GLUTAMIC ACID, Glutaminase kidney isoform, mitochondrial | | Authors: | DeLaBarre, B, Gross, S, Cheng, F, Gao, Y, Jha, A, Jiang, F, Song, J.J, Wie, W, Hurov, J. | | Deposit date: | 2011-11-16 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Full-length human glutaminase in complex with an allosteric inhibitor.
Biochemistry, 50, 2011
|
|
