4KQO
 
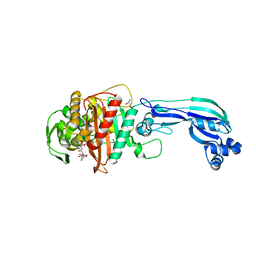 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4IV1
 
 | | Crystal structure of recombinant foot-and-mouth-disease virus A22 empty capsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Porta, C, Kotecha, A, Burman, A, Jackson, T, Ren, J, Loureiro, S, Jones, I.M, Fry, E.E, Stuart, D.I, Charleston, B. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen.
Plos Pathog., 9, 2013
|
|
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4IV3
 
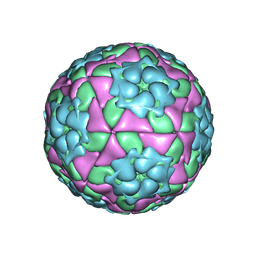 | | Crystal structure of recombinant foot-and-mouth-disease virus A22-H2093C empty capsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Porta, C, Kotecha, A, Burman, A, Jackson, T, Ren, J, Loureiro, S, Jones, I.M, Fry, E.E, Stuart, D.I, Charleston, B. | | Deposit date: | 2013-01-22 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational engineering of recombinant picornavirus capsids to produce safe, protective vaccine antigen.
Plos Pathog., 9, 2013
|
|
8AYY
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8AYX
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8AYZ
 
 | | Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17 | | Descriptor: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.88 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
3HHF
 
 | | Structure of CrgA regulatory domain, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
6Z3P
 
 | | Structure of EV71 in complex with a protective antibody 38-3-11A Fab | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
3HHG
 
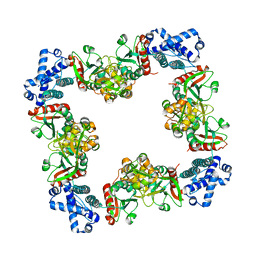 | | Structure of CrgA, a LysR-type transcriptional regulator from Neisseria meningitidis. | | Descriptor: | Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Owens, R.J, Stuart, D.I, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-05-15 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators
Nucleic Acids Res., 37, 2009
|
|
6Z3Q
 
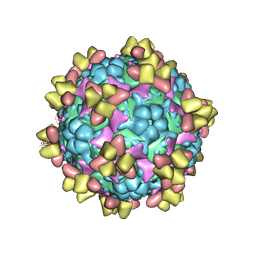 | | Structure of EV71 in complex with a protective antibody 38-1-10A Fab | | Descriptor: | Heavy chain, Light chain, SPHINGOSINE, ... | | Authors: | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-05-21 | | Release date: | 2020-09-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
6Z6W
 
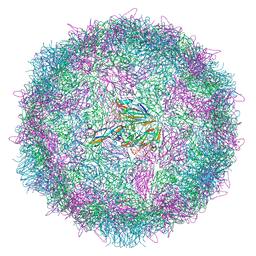 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) from a mammalian expression system. | | Descriptor: | Capsid proteins, VP0, VP1, ... | | Authors: | Bahar, M.W, Porta, C, Fry, E.E, Stuart, D.I. | | Deposit date: | 2020-05-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mammalian expression of virus-like particles as a proof of principle for next generation polio vaccines.
NPJ Vaccines, 6, 2021
|
|
6ZFO
 
 | | Association of two complexes of largely structurally disordered Spike ectodomain with bound EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, EY6A light chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6Z3K
 
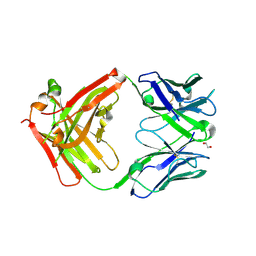 | | Structure of protective antibody 38-1-10A Fab | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain, Light Chain | | Authors: | Zhou, D, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-05-20 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of protective antibodies targeting the threefold plateau of enterovirus 71.
Nat Commun, 11, 2020
|
|
6Z97
 
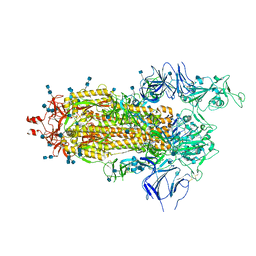 | | Structure of the prefusion SARS-CoV-2 spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Duyvesteyn, H.M.E, Ren, J, Zhao, Y, Zhou, D, Huo, J, Carrique, L, Malinauskas, T, Ruza, R.R, Shah, P.N.M, Fry, E.E, Owens, R, Stuart, D.I. | | Deposit date: | 2020-06-03 | | Release date: | 2020-07-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.
Cell Host Microbe, 28, 2020
|
|
6ZDH
 
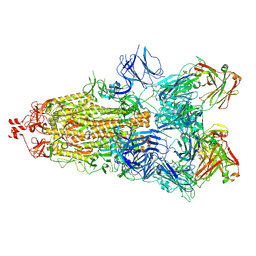 | | SARS-CoV-2 Spike glycoprotein in complex with a neutralizing antibody EY6A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EY6A heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Zhou, D, Zhao, Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2020-06-14 | | Release date: | 2020-07-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6J7V
 
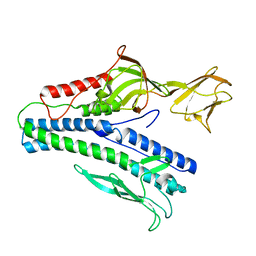 | |
6HS4
 
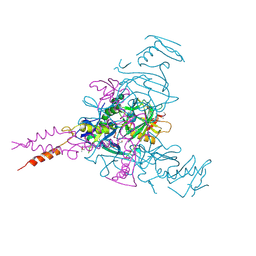 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118 | | Descriptor: | 1-[2-[3-oxidanyl-4-(4-phenyl-1~{H}-pyrazol-5-yl)phenoxy]ethyl]piperidine-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Stuart, D.I. | | Deposit date: | 2018-09-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
6HY0
 
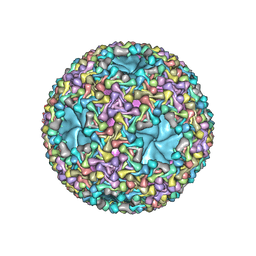 | | Atomic models of P1, P4 C-terminal fragment and P8 fitted in the bacteriophage phi6 nucleocapsid reconstructed with icosahedral symmetry | | Descriptor: | Major Outer Capsid Protein P8, Major inner protein P1, Packaging Enzyme P4 | | Authors: | El Omari, K, Ilca, S.L, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2018-10-18 | | Release date: | 2019-06-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multiple liquid crystalline geometries of highly compacted nucleic acid in a dsRNA virus.
Nature, 570, 2019
|
|
6HRO
 
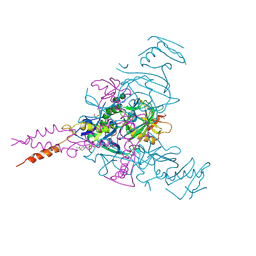 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118a | | Descriptor: | 1-[2-[4-[4-(4-chlorophenyl)-3-methyl-1~{H}-pyrazol-5-yl]-3-oxidanyl-phenoxy]ethyl]piperidin-1-ium-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Stuart, D.I. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
6I2K
 
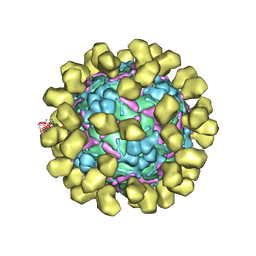 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
3JV9
 
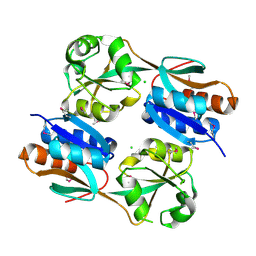 | | The structure of a reduced form of OxyR from N. meningitidis | | Descriptor: | CHLORIDE ION, Transcriptional regulator, LysR family | | Authors: | Sainsbury, S, Ren, J, Stuart, D.I, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-09-16 | | Release date: | 2010-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The structure of a reduced form of OxyR from Neisseria meningitidis
Bmc Struct.Biol., 10, 2010
|
|
2VHJ
 
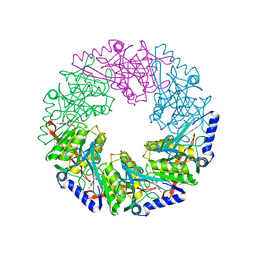 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHT
 
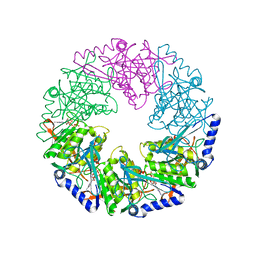 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 R279A mutant in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-25 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
5JQ7
 
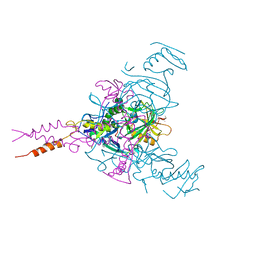 | | Crystal structure of Ebola glycoprotein in complex with toremifene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Envelope glycoprotein 1,Envelope glycoprotein 1,Envelope glycoprotein 1, ... | | Authors: | Zhao, Y, Ren, J, Stuart, D.I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Toremifene interacts with and destabilizes the Ebola virus glycoprotein.
Nature, 535, 2016
|
|
