1RYF
 
 | |
1NZB
 
 | | Crystal structure of wild type Cre recombinase-loxP synapse | | Descriptor: | Cre recombinase, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Meyer, J.E.W, Buchholz, F, Stewart, A.F, Suck, D. | | Deposit date: | 2003-02-17 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of a wild-type Cre recombinase-loxP synapse reveals a novel spacer conformation suggesting an alternative mechanism for DNA cleavage activation
Nucleic Acids Res., 31, 2003
|
|
5KYK
 
 | | Covalent GTP-competitive inhibitors of KRAS G12C: Guanosine bisphosphonate Analogs | | Descriptor: | 5'-O-[(R)-[({2-[(chloroacetyl)amino]ethyl}sulfamoyl)methyl](hydroxy)phosphoryl]guanosine, GTPase KRas | | Authors: | Xiong, Y, Lu, J, Hunter, J, Li, L, Scott, D, Manandhar, A, Gondi, S, Westover, K.D, Gray, N.S. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Covalent Guanosine Mimetic Inhibitors of G12C KRAS.
ACS Med Chem Lett, 8, 2017
|
|
1NLM
 
 | | CRYSTAL STRUCTURE OF MURG:GLCNAC COMPLEX | | Descriptor: | GLYCEROL, UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Hu, Y, Chen, L, Ha, S, Gross, B, Falcone, B, Walker, D, Mokhtarzadeh, M, Walker, S. | | Deposit date: | 2003-01-07 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of MurG:UDP-GlcNAc complex reveals common structural principles of a superfamily of glycosyltransferases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1S0B
 
 | | Crystal structure of botulinum neurotoxin type B at pH 4.0 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
1S0Z
 
 | | Crystal structure of the VDR LBD complexed to seocalcitol. | | Descriptor: | SEOCALCITOL, Vitamin D3 receptor | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Moras, D. | | Deposit date: | 2004-01-05 | | Release date: | 2004-04-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the vitamin D nuclear receptor liganded with the vitamin D side chain analogues calcipotriol and seocalcitol, receptor agonists of clinical importance. Insights into a structural basis for the switching of calcipotriol to a receptor antagonist by further side chain modification.
J.Med.Chem., 47, 2004
|
|
7NQC
 
 | | Calmodulin extracts the Ras family protein RalA from lipid bilayers by engagement with two membrane targeting motifs | | Descriptor: | CALCIUM ION, Calmodulin-1, PRO-ASN-GLY-LYS-LYS-LYS-ARG-LYS-SER-LEU-ALA-LYS-ARG-ILE-ARG-GLU-ARG-CMF, ... | | Authors: | Chamberlain, S.G, Owen, D, Mott, H.R. | | Deposit date: | 2021-03-01 | | Release date: | 2021-09-22 | | Method: | SOLUTION NMR | | Cite: | Calmodulin extracts the Ras family protein RalA from lipid bilayers by engagement with two membrane-targeting motifs.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1RYG
 
 | |
1NM8
 
 | | Structure of Human Carnitine Acetyltransferase: Molecular Basis for Fatty Acyl Transfer | | Descriptor: | Carnitine O-acetyltransferase | | Authors: | Wu, D, Govindasamy, L, Lian, W, Gu, Y, Kukar, T, Agbandje-McKenna, M, McKenna, R. | | Deposit date: | 2003-01-09 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Human Carnitine Acetyltransferase. Molecular Basis for Fatty Acyl Transfer
J.Biol.Chem., 278, 2003
|
|
7NXV
 
 | | Crystal structure of the complex of DNase I/G-actin/PPP1R15A_582-621 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yan, Y, Ron, D. | | Deposit date: | 2021-03-19 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Higher-order phosphatase-substrate contacts terminate the integrated stress response.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7NZM
 
 | | Cryo-EM structure of pre-dephosphorylation complex of phosphorylated eIF2alpha with trapped holophosphatase (PP1A_D64A/PPP1R15A/G-actin/DNase I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yan, Y, Hardwick, S, Ron, D. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Higher-order phosphatase-substrate contacts terminate the integrated stress response.
Nat.Struct.Mol.Biol., 28, 2021
|
|
2RIN
 
 | | ABC-transporter choline binding protein in complex with acetylcholine | | Descriptor: | ACETYLCHOLINE, PUTATIVE GLYCINE BETAINE-BINDING ABC TRANSPORTER PROTEIN | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Sohn-Boeser, L, Le Rudulier, D, Schmitt, L, Bremer, E. | | Deposit date: | 2007-10-12 | | Release date: | 2008-09-30 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the choline/acetylcholine substrate-binding protein ChoX from Sinorhizobium meliloti in the liganded and unliganded-closed states.
J.Biol.Chem., 283, 2008
|
|
1NOU
 
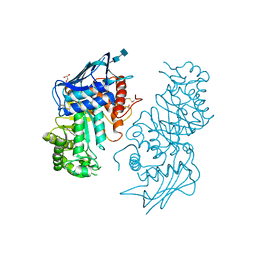 | | Native human lysosomal beta-hexosaminidase isoform B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
5L6Z
 
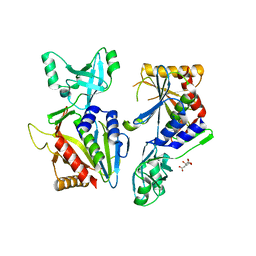 | |
5KKZ
 
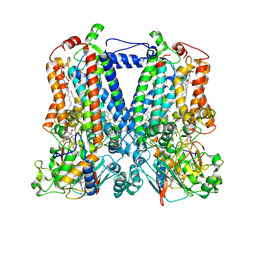 | | Rhodobacter sphaeroides bc1 with famoxadone | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, ASCORBIC ACID, Cytochrome b, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Tang, W.K, Yu, C.A. | | Deposit date: | 2016-06-23 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
1S3Q
 
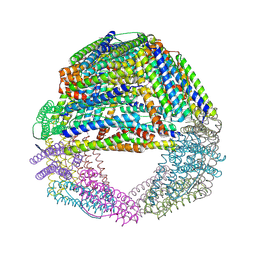 | | Crystal structures of a novel open pore ferritin from the hyperthermophilic Archaeon Archaeoglobus fulgidus | | Descriptor: | ZINC ION, ferritin | | Authors: | Johnson, E, Cascio, D, Sawaya, M, Schroeder, I. | | Deposit date: | 2004-01-13 | | Release date: | 2005-04-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of a tetrahedral open pore ferritin from the hyperthermophilic archaeon Archaeoglobus fulgidus.
Structure, 13, 2005
|
|
1NQ3
 
 | | Crystal structure of the mammalian tumor associated antigen UK114 | | Descriptor: | 14.3 kDa perchloric acid soluble protein | | Authors: | Deriu, D, Briand, C, Mistiniene, E, Naktinis, V, Grutter, M.G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and oligomeric state of the mammalian tumour-associated antigen UK114.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
5KNP
 
 | | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with [3S,4R]-(4-(Hypoxanthin-9-yl)pyrrolidin-3-yl)-oxymethanephosphonic acid | | Descriptor: | Hypoxanthine-guanine phosphoribosyltransferase, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Eng, W.S, Rejman, D, Keough, D.T, Guddat, L.W. | | Deposit date: | 2016-06-28 | | Release date: | 2017-09-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis hypoxanthine guanine phosphoribosyltransferase in complex with pyrrolidine nucleoside phosphonate
To Be Published
|
|
7NYC
 
 | | cryoEM structure of 3C9-sMAC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Menny, A, Couves, E.C, Bubeck, D. | | Deposit date: | 2021-03-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
1S5G
 
 | | Structure of Scallop myosin S1 reveals a novel nucleotide conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Risal, D, Gourinath, S, Himmel, D.M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2004-01-20 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S0D
 
 | | Crystal structure of botulinum neurotoxin type B at pH 5.5 | | Descriptor: | Botulinum neurotoxin type B, CALCIUM ION, ZINC ION | | Authors: | Eswaramoorthy, S, Kumaran, D, Keller, J, Swaminathan, S. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of metals in the biological activity of Clostridium botulinum neurotoxins
Biochemistry, 43, 2004
|
|
5L86
 
 | | engineered ascorbate peroxidise | | Descriptor: | Ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hayashi, T, Mittl, P, Hilvert, D. | | Deposit date: | 2016-06-07 | | Release date: | 2017-03-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Chemically Programmed Proximal Ligand Enhances the Catalytic Properties of a Heme Enzyme.
J. Am. Chem. Soc., 138, 2016
|
|
1NTZ
 
 | | Crystal Structure of Mitochondrial Cytochrome bc1 Complex Bound with Ubiquinone | | Descriptor: | Cytochrome b, FE2/S2 (INORGANIC) CLUSTER, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Gao, X, Wen, X, Esser, L, Quinn, B, Yu, L, Yu, C.-A, Xia, D. | | Deposit date: | 2003-01-30 | | Release date: | 2003-10-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the quinone reduction in the bc(1) complex: a comparative analysis of crystal structures of mitochondrial cytochrome bc(1) with bound substrate and inhibitors at the Q(i) site
Biochemistry, 42, 2003
|
|
2ROE
 
 | | Solution structure of thermus thermophilus HB8 TTHA1718 protein in vitro | | Descriptor: | Heavy metal binding protein | | Authors: | Sakakibara, D, Sasaki, A, Ikeya, T, Hamatsu, J, Koyama, H, Mishima, M, Mikawa, T, Waelchli, M, Smith, B.O, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2008-03-20 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein structure determination in living cells by in-cell NMR spectroscopy
Nature, 458, 2009
|
|
1S5N
 
 | | Xylose Isomerase in Substrate and Inhibitor Michaelis States: Atomic Resolution Studies of a Metal-Mediated Hydride Shift | | Descriptor: | HYDROXIDE ION, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Fenn, T.D, Ringe, D, Petsko, G.A. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Xylose isomerase in substrate and inhibitor michaelis States: atomic resolution studies of a metal-mediated hydride shift(,).
Biochemistry, 43, 2004
|
|
