7DCC
 
 | |
5XDM
 
 | |
5ZHE
 
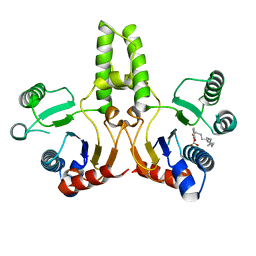 | | STRUCTURE OF E. COLI UNDECAPRENYL DIPHOSPHATE SYNTHASE IN COMPLEX WITH BPH-981 | | Descriptor: | 2-hydroxy-6-(tetradecyloxy)benzoic acid, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
7WWM
 
 | | S protein of Delta variant in complex with ZWC6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7WWL
 
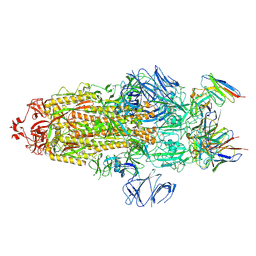 | | S protein of Delta variant in complex with ZWD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-13 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination.
Signal Transduct Target Ther, 7, 2022
|
|
7XC2
 
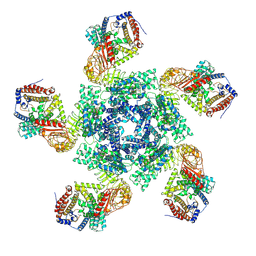 | | Cryo EM structure of oligomeric complex formed by wheat CNL Sr35 and the effector AvrSr35 of the wheat stem rust pathogen | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Avirulence factor, CNL9 | | Authors: | Alexander, F, Li, E.T, Aaron, L, Deng, Y.N, Sun, Y, Chai, J.J. | | Deposit date: | 2022-03-22 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A wheat resistosome defines common principles of immune receptor channels.
Nature, 610, 2022
|
|
1UEF
 
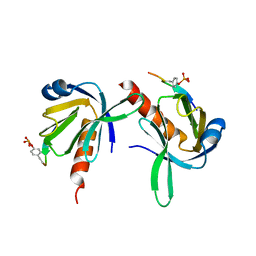 | | Crystal Structure of Dok1 PTB Domain Complex | | Descriptor: | 13-mer peptide from Proto-oncogene tyrosine-protein kinase receptor ret, Docking protein 1 | | Authors: | Shi, N, Ye, S, Liu, Y, Zhou, W, Ding, Y, Lou, Z, Qiang, B, Yan, J, Rao, Z. | | Deposit date: | 2003-05-14 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Specific Recognition of RET by the Dok1 Phosphotyrosine Binding Domain
J.Biol.Chem., 279, 2004
|
|
7V6F
 
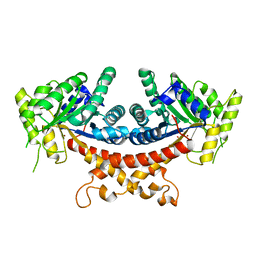 | | Structure of Candida albicans Fructose-1,6-bisphosphate aldolase complexed with G3P | | Descriptor: | Fructose-bisphosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ZINC ION | | Authors: | Hongxuan, C, Huang, Y, Han, C, Chen, W, Ren, Y, Wan, J. | | Deposit date: | 2021-08-20 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-Guided Discovery of the Novel Covalent Allosteric Site and Covalent Inhibitors of Fructose-1,6-Bisphosphate Aldolase to Overcome the Azole Resistance of Candidiasis.
J.Med.Chem., 65, 2022
|
|
4UTB
 
 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UT7
 
 | | CRYSTAL STRUCTURE OF THE SCFV FRAGMENT OF THE BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
7CRW
 
 | | Cryo-EM structure of rNLRP1-rDPP9 complex | | Descriptor: | Dipeptidyl peptidase 9, NLR family protein 1 | | Authors: | Huang, M.H, Zhang, X.X, Wang, J, Chai, J.J. | | Deposit date: | 2020-08-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural and biochemical mechanisms of NLRP1 inhibition by DPP9.
Nature, 592, 2021
|
|
7CRV
 
 | |
6A5F
 
 | |
6A5H
 
 | |
6TI2
 
 | |
6A5G
 
 | |
6VN2
 
 | | USP7 IN COMPLEX WITH LIGAND COMPOUND 18 | | Descriptor: | 1-({7-[(2R)-5-chloro-2-(piperazine-1-carbonyl)-2,3-dihydro-1-benzofuran-7-yl]thieno[3,2-b]pyridin-2-yl}methyl)-1H-pyrrole-2,5-dione, ACETATE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Leger, P.R, Wustrow, D.J, Hu, D.X, Krapp, S, Maskos, K, Blaesse, M. | | Deposit date: | 2020-01-29 | | Release date: | 2020-04-29 | | Last modified: | 2020-06-17 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of Potent, Selective, and Orally Bioavailable Inhibitors of USP7 with In Vivo Antitumor Activity.
J.Med.Chem., 63, 2020
|
|
6A6J
 
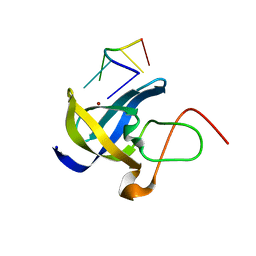 | | Crystal structure of Zebra fish Y-box protein1 (YB-1) Cold-shock domain in complex with 6mer m5C RNA | | Descriptor: | RNA (5'-R(P*CP*AP*UP*(5MC)P*U)-3'), ZINC ION, Zebra fish Y-box protein1 (YB-1) | | Authors: | Zhang, M.M, Wu, B.X, Huang, Y, Ma, J.B. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay.
Mol.Cell, 75, 2019
|
|
7SL2
 
 | |
7SL6
 
 | |
7SL3
 
 | |
7SL4
 
 | |
7SL7
 
 | |
7SL1
 
 | |
7SBY
 
 | | Structure of OC43 spike in complex with polyclonal Fab7 (Donor 269) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Human polyclonal Fab model with polyalanine backbone - Heavy chain, ... | | Authors: | Bangaru, S, Antanasijevic, A, Ward, A. | | Deposit date: | 2021-09-26 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mapping of antibody landscapes to human betacoronavirus spike proteins.
Sci Adv, 8, 2022
|
|
