1Q9U
 
 | |
3N67
 
 | | Structure of endothelial nitric oxide synthase N368D/V106M double mutant heme domain complexed with 6,6'-(2,2'-(5-amino-1,3-phenylene)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-[(5-aminobenzene-1,3-diyl)diethane-2,1-diyl]bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
3N5Q
 
 | | Structure of endothelial nitric oxide synthase heme domain complexed with 6,6'-(2,2'-(5-amino-1,3-phenylene)bis(ethane-2,1-diyl))bis(4-methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-[(5-aminobenzene-1,3-diyl)diethane-2,1-diyl]bis(4-methylpyridin-2-amine), CACODYLIC ACID, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2010-05-25 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Role of zinc in isoform-selective inhibitor binding to neuronal nitric oxide synthase .
Biochemistry, 49, 2010
|
|
1Q1W
 
 | |
1PKH
 
 | | STRUCTURAL BASIS FOR RECOGNITION AND CATALYSIS BY THE BIFUNCTIONAL DCTP DEAMINASE AND DUTPASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional deaminase/diphosphatase | | Authors: | Huffman, J.L, Li, H, White, R.H, Tainer, J.A. | | Deposit date: | 2003-06-05 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii
J.Mol.Biol., 331, 2003
|
|
7E74
 
 | |
5EID
 
 | | AKR2A ankyrin repeat domain | | Descriptor: | Ankyrin repeat domain-containing protein 2 | | Authors: | Pu, H, Li, H. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | structure of AKR2A ankyrin repeat domain at 2.0 angstroms resolution
To Be Published
|
|
1JFF
 
 | | Refined structure of alpha-beta tubulin from zinc-induced sheets stabilized with taxol | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lowe, J, Li, H, Downing, K.H, Nogales, E. | | Deposit date: | 2001-06-20 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.5 Å) | | Cite: | Refined structure of alpha beta-tubulin at 3.5 A resolution.
J.Mol.Biol., 313, 2001
|
|
1JCI
 
 | | Stabilization of the Engineered Cation-binding Loop in Cytochrome c Peroxidase (CcP) | | Descriptor: | Cytochrome C Peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhaskar, B, Bonagura, C.A, Li, H, Poulos, T.L. | | Deposit date: | 2001-06-09 | | Release date: | 2002-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cation-induced stabilization of the engineered cation-binding loop in cytochrome c peroxidase (CcP).
Biochemistry, 41, 2002
|
|
4APP
 
 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with (S)-N-(5-(3-benzyl-1-methylpiperazine-4-carbonyl)-6,6-dimethyl-1,4,5, 6-tetrahydropyrrolo(3,4-c)pyrazol-3-yl)-3-phenoxybenzamide | | Descriptor: | GLYCEROL, N-[6,6-dimethyl-5-[(2S)-4-methyl-2-(phenylmethyl)piperazin-1-yl]carbonyl-2,4-dihydropyrrolo[3,4-c]pyrazol-3-yl]-3-phenoxy-benzamide, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.D, Deng, Y.L, Wang, C, Guo, C, McAlpine, I, Zhang, J, Kephart, S, Johnson, M.C, Li, H, Bouzida, D, Yang, A, Dong, L, Marakovits, J, Tikhe, J, Richardson, P, Guo, L.C, Kania, R, Edwards, M.P, Kraynov, E, Christensen, J, Piraino, J, Lee, J, Dagostino, E, Del-Carmen, C, Smeal, T, Murray, B.W. | | Deposit date: | 2012-04-04 | | Release date: | 2012-06-06 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Pyrroloaminopyrazoles as Novel Pak Inhibitors.
J.Med.Chem., 55, 2012
|
|
4AZN
 
 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag-mediated crystal packing | | Descriptor: | FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1KTK
 
 | | Complex of Streptococcal pyrogenic enterotoxin C (SpeC) with a human T cell receptor beta chain (Vbeta2.1) | | Descriptor: | Exotoxin type C, T-cell receptor beta chain | | Authors: | Sundberg, E.J, Li, H, Llera, A.S, McCormick, J.K, Tormo, J, Karjalainen, K, Schlievert, P.M, Mariuzza, R.A. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of two streptococcal superantigens bound to TCR beta chains reveal diversity in the architecture of T cell signaling complexes.
Structure, 10, 2002
|
|
4AZP
 
 | | Murine epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid anandamide | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZR
 
 | | Human epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid anandamide | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1KFO
 
 | | CRYSTAL STRUCTURE OF AN RNA HELIX RECOGNIZED BY A ZINC-FINGER PROTEIN: AN 18 BASE PAIR DUPLEX AT 1.6 RESOLUTION | | Descriptor: | 5'-R(*GP*AP*AP*UP*GP*CP*CP*UP*GP*CP*GP*AP*GP*CP*AP*(5BU)P*CP*CP*C)-3' | | Authors: | Lima, S, Hildenbrand, J, Korostelev, A, Hattman, S, Li, H. | | Deposit date: | 2001-11-21 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of an RNA helix recognized by a zinc-finger protein: an 18-bp duplex at 1.6 A resolution.
RNA, 8, 2002
|
|
4ZDN
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsF KS4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AT-less polyketide synthase, CHLORIDE ION | | Authors: | Chang, C, Li, H, Endres, M, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-04-17 | | Release date: | 2015-05-13 | | Last modified: | 2023-03-22 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4AZO
 
 | | Murine epidermal fatty acid-binding protein (FABP5), apo form, poly- his tag removed | | Descriptor: | CHLORIDE ION, FATTY ACID-BINDING PROTEIN, EPIDERMAL | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZQ
 
 | | Murine epidermal fatty acid-binding protein (FABP5) in complex with the endocannabinoid 2-arachidonoylglycerol | | Descriptor: | 2-hydroxy-1-(hydroxymethyl)ethyl icosanoate, CHLORIDE ION, FATTY ACID-BINDING PROTEIN, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4AZM
 
 | | Human epidermal fatty acid-binding protein (FABP5) in complex with the inhibitor BMS-309413 | | Descriptor: | ((2'-(5-ETHYL-3,4-DIPHENYL-1H-PYRAZOL-1-YL)-3-BIPHENYLYL)OXY)ACETIC ACID, FATTY ACID-BINDING PROTEIN, EPIDERMAL, ... | | Authors: | Sanson, B, Wang, T, Sun, J, Kaczocha, M, Ojima, I, Deutsch, D, Li, H. | | Deposit date: | 2012-06-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystallographic Study of Fabp5 as an Intracellular Endocannabinoid Transporter.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4ZR7
 
 | | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168 | | Descriptor: | ACETATE ION, CHLORIDE ION, Sensor histidine kinase ResE | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-11 | | Release date: | 2015-05-27 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of a domain of a functionally unknown protein from Bacillus subtilis subsp. subtilis str. 168
To Be Published
|
|
3B3P
 
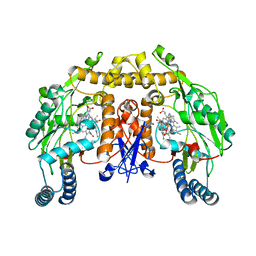 | | Structure of neuronal nos heme domain in complex with a inhibitor (+-)-n1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-n2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4S)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-(3-chlorobenzyl)ethane-1,2-diamine, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2007-10-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
1MMW
 
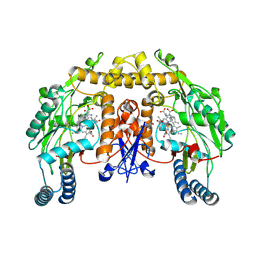 | | Rat neuronal NOS heme domain with vinyl-L-NIO bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N5-(1-IMINO-3-BUTENYL)-L-ORNITHINE, ... | | Authors: | Bretscher, L.E, Li, H, Poulos, T.L, Griffith, O.W. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization and kinetics of nitric-oxide synthase inhibition by novel N5-(iminoalkyl)- and N5-(iminoalkenyl)-ornithines
J.Biol.Chem., 278, 2003
|
|
1MMV
 
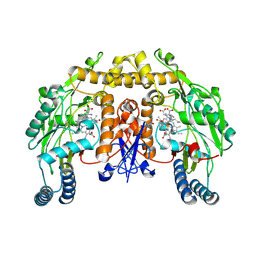 | | Rat neuronal NOS heme domain with NG-propyl-L-arginine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-PROPYL-L-ARGININE, ... | | Authors: | Bretscher, L.E, Li, H, Poulos, T.L, Griffith, O.W. | | Deposit date: | 2002-09-04 | | Release date: | 2003-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization and kinetics of nitric-oxide synthase inhibition by novel N5-(iminoalkyl)- and N5-(iminoalkenyl)-ornithines
J.Biol.Chem., 278, 2003
|
|
3QJJ
 
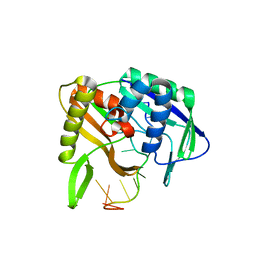 | |
3QJP
 
 | |
