4OHE
 
 | | LEOPARD Syndrome-Associated SHP2/G464A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
4OHI
 
 | | LEOPARD Syndrome-Associated SHP2/Q510E mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Yu, Z.H, Zhang, R.Y, Walls, C.D, Chen, L, Zhang, S, Wu, L, Wang, L, Liu, S, Zhang, Z.Y. | | Deposit date: | 2014-01-17 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations.
Biochemistry, 53, 2014
|
|
7D99
 
 | | human potassium-chloride co-transporter KCC4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
5E38
 
 | | Structural basis of mapping the spontaneous mutations with 5-flourouracil in uracil phosphoribosyltransferase from Mycobacterium tuberculosis | | Descriptor: | Uracil phosphoribosyltransferase | | Authors: | Ghode, P, Jobichen, C, Ramachandran, S, Bifani, P, Sivaraman, J. | | Deposit date: | 2015-10-02 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of mapping the spontaneous mutations with 5-flurouracil in uracil phosphoribosyltransferase from Mycobacterium tuberculosis
Biochem.Biophys.Res.Commun., 467, 2015
|
|
1I4P
 
 | |
7D90
 
 | | human potassium-chloride co-transporter KCC3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 3 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-12 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
7D8Z
 
 | | human potassium-chloride co-transporter KCC2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, potassium-chloride cotransporter 2 | | Authors: | Xie, Y, Chang, S, Zhao, C, Ye, S, Guo, J. | | Deposit date: | 2020-10-11 | | Release date: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and an activation mechanism of human potassium-chloride cotransporters.
Sci Adv, 6, 2020
|
|
3R6A
 
 | |
5IAO
 
 | |
2I3O
 
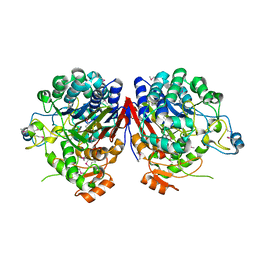 | |
3EBN
 
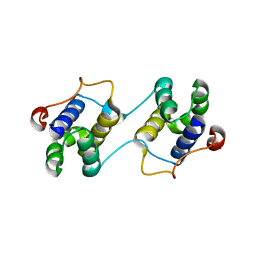 | | A Special Dimerization of SARS-CoV Main Protease C-Terminal Domain Due to Domain-swapping | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Zhong, N, Zhang, S, Xue, F, Kang, X, Lou, Z, Xia, B. | | Deposit date: | 2008-08-28 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | C-terminal domain of SARS-CoV main protease can form a 3D domain-swapped dimer
PROTEIN SCI., 18, 2009
|
|
2IMR
 
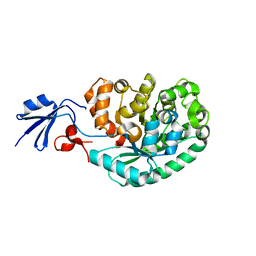 | |
5X8R
 
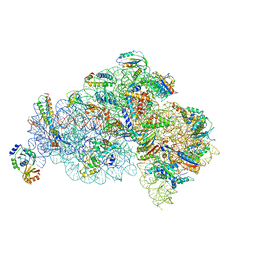 | | Structure of the 30S small subunit of chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 30S ribosomal protein S1, chloroplastic, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
4HL9
 
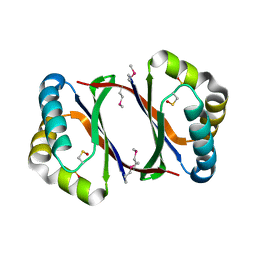 | | Crystal structure of antibiotic biosynthesis monooxygenase | | Descriptor: | Antibiotic biosynthesis monooxygenase | | Authors: | Rice, S, Eswaramoorthy, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-10-31 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of antibiotic biosynthesis monooxygenase
To be Published
|
|
5X8P
 
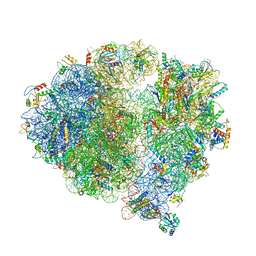 | | Structure of the 70S chloroplast ribosome from spinach | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
2EA1
 
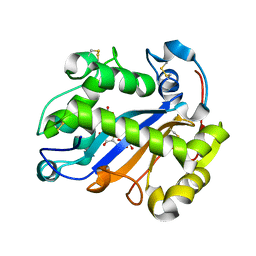 | | Crystal structure of Ribonuclease I from Escherichia coli COMPLEXED WITH GUANYLYL-2(PRIME),5(PRIME)-GUANOSINE | | Descriptor: | GUANYLYL-2',5'-PHOSPHOGUANOSINE, Ribonuclease I | | Authors: | Zhou, K, Pan, J, Padmanabhan, S, Lim, R.W, Lim, L.W. | | Deposit date: | 2007-01-29 | | Release date: | 2008-01-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Ribonuclease I from Escherichia Coli Complexed with Guanylyl-2(Prime),5(Prime)-Guanosine at 1.80 Angstroms Resolution
To be Published
|
|
5X8T
 
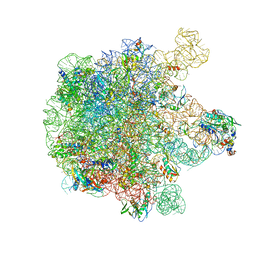 | | Structure of the 50S large subunit of chloroplast ribosome from spinach | | Descriptor: | 23S rRNA, 4.8S rRNA, 50S ribosomal protein L13, ... | | Authors: | Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Unique localization of the plastid-specific ribosomal proteins in the chloroplast ribosome small subunit provides mechanistic insights into the chloroplastic translation
Nucleic Acids Res., 45, 2017
|
|
7WF3
 
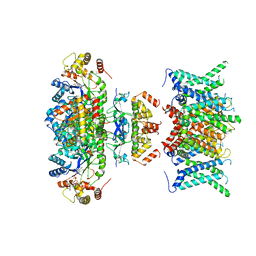 | | Composite map of human Kv1.3 channel in apo state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7WF4
 
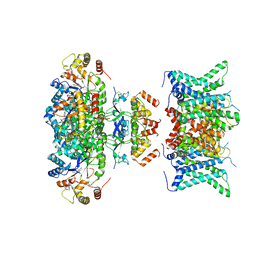 | | Composite map of human Kv1.3 channel in dalazatide-bound state with beta subunits | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 3, ... | | Authors: | Tyagi, A, Ahmed, T, Jian, S, Bajaj, S, Ong, S.T, Goay, S.S.M, Zhao, Y, Vorobyov, I, Tian, C, Chandy, K.G, Bhushan, S. | | Deposit date: | 2021-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rearrangement of a unique Kv1.3 selectivity filter conformation upon binding of a drug.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5FSW
 
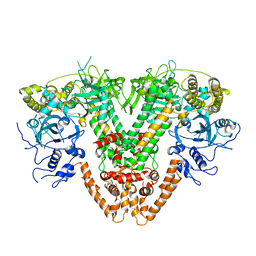 | | RNA dependent RNA polymerase QDE-1 from Thielavia terrestris | | Descriptor: | RNA DEPENDENT RNA POLYMERASE QDE-1 | | Authors: | Qian, X, Hamid, F.M, El Sahili, A, Darwis, D.A, Wong, Y.H, Bhushan, S, Makeyev, E.V, Lescar, J. | | Deposit date: | 2016-01-08 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Functional Evolution in Orthologous Cell-Encoded RNA-Dependent RNA Polymerases
J.Biol.Chem., 291, 2016
|
|
1TXZ
 
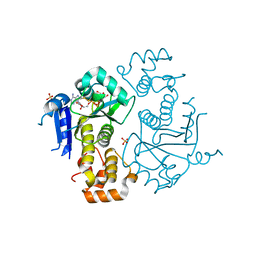 | | Crystal structure of yeast ymx7, an ADP-ribose-1''-monophosphatase, complexed with ADP-ribose | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5-DIPHOSPHORIBOSE, Hypothetical 32.1 kDa protein in ADH3-RCA1 intergenic region, ... | | Authors: | Kumaran, D, Swaminathan, S, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-07-06 | | Release date: | 2004-11-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and mechanism of ADP-ribose-1''-monophosphatase (Appr-1''-pase), a ubiquitous cellular processing enzyme.
Protein Sci., 14, 2005
|
|
3R79
 
 | |
2I76
 
 | | Crystal structure of protein TM1727 from Thermotoga maritima | | Descriptor: | Hypothetical protein, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Madegowda, M, Eswaramoorthy, S, Seetharaman, J, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-08-30 | | Release date: | 2006-10-03 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of hypothetical protein TM1727 from Thermatoga maritima
TO BE PUBLISHED
|
|
4FGS
 
 | | Crystal structure of a probable dehydrogenase protein | | Descriptor: | Probable dehydrogenase protein, SULFATE ION | | Authors: | Eswaramoorthy, S, Rice, S, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of a probable dehydrogenase protein
To be Published
|
|
3T8Q
 
 | | Crystal structure of mandelate racemase/muconate lactonizing enzyme family protein from Hoeflea phototrophica | | Descriptor: | MAGNESIUM ION, MALONATE ION, Mandelate racemase/muconate lactonizing enzyme family protein | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mandelate racemase/muconate lactonizing enzyme family protein from Hoeflea phototrophica
To be Published
|
|
