1AT9
 
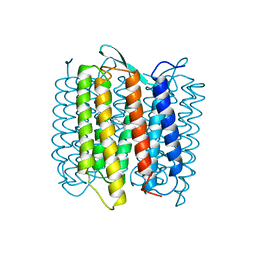 | | STRUCTURE OF BACTERIORHODOPSIN AT 3.0 ANGSTROM DETERMINED BY ELECTRON CRYSTALLOGRAPHY | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Kimura, Y, Vassylyev, D.G, Miyazawa, A, Kidera, A, Matsushima, M, Mitsuoka, K, Murata, K, Hirai, T, Fujiyoshi, Y. | | Deposit date: | 1997-08-20 | | Release date: | 1998-09-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Surface of bacteriorhodopsin revealed by high-resolution electron crystallography.
Nature, 389, 1997
|
|
2LPU
 
 | | Solution structures of KmAtg10 | | Descriptor: | KmAtg10 | | Authors: | Yamaguchi, M, Noda, N.N, Yamamoto, H, Shima, T, Kumeta, H, Kobashigawa, Y, Akada, R, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2012-02-19 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into atg10-mediated formation of the autophagy-essential atg12-atg5 conjugate
Structure, 20, 2012
|
|
3W39
 
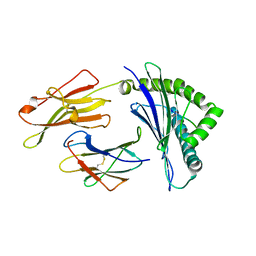 | | Crystal structure of HLA-B*5201 in complexed with HIV immunodominant epitope (TAFTIPSI) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-52 alpha chain, ... | | Authors: | Yagita, Y, Kuse, N, Kuroki, K, Gatanaga, H, Carlson, J.M, Chikata, T, Brumme, Z.L, Murakoshi, H, Akahoshi, T, Pfeifer, N, Mallal, S, John, M, Ose, T, Matsubara, H, Kanda, R, Fukunaga, Y, Honda, K, Kawashima, Y, Ariumi, Y, Oka, S, Maenaka, K, Takiguchi, M. | | Deposit date: | 2012-12-13 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct HIV-1 Escape Patterns Selected by Cytotoxic T Cells with Identical Epitope Specificity
J.Virol., 87, 2013
|
|
4IJ6
 
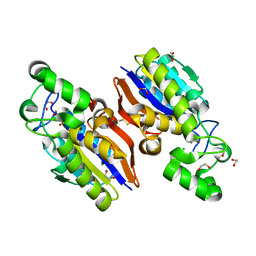 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase Mutant (H9A) from Hydrogenobacter thermophilus TK-6 in Complex with L-phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHOSERINE, ... | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
3IYZ
 
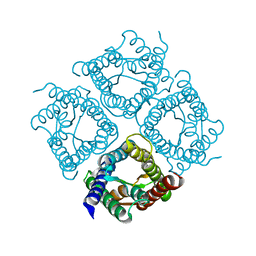 | | Structure of Aquaporin-4 S180D mutant at 10.0 A resolution from electron micrograph | | Descriptor: | Aquaporin-4 | | Authors: | Mitsuma, T, Tani, K, Hiroaki, Y, Kamegawa, A, Suzuki, H, Hibino, H, Kurachi, Y, Fujiyoshi, Y. | | Deposit date: | 2010-07-24 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (10 Å) | | Cite: | Influence of the cytoplasmic domains of aquaporin-4 on water conduction and array formation.
J.Mol.Biol., 402, 2010
|
|
1CL6
 
 | |
1UIL
 
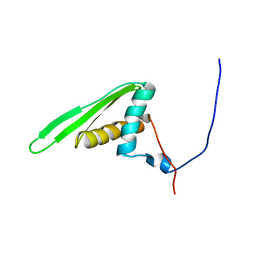 | | Double-stranded RNA-binding motif of Hypothetical protein BAB28848 | | Descriptor: | Double-stranded RNA-binding motif | | Authors: | Nagata, T, Muto, Y, Hayashi, F, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-17 | | Release date: | 2004-11-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Double-stranded RNA-binding motif of Hypothetical protein BAB28848
To be Published
|
|
1CMN
 
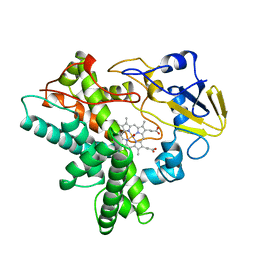 | |
1AN9
 
 | | D-AMINO ACID OXIDASE COMPLEX WITH O-AMINOBENZOATE | | Descriptor: | 2-AMINOBENZOIC ACID, D-AMINO ACID OXIDASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Miura, R, Setoyama, C, Nishina, Y, Shiga, K, Mizutani, H, Miyahara, I, Hirotsu, K. | | Deposit date: | 1997-06-28 | | Release date: | 1997-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and mechanistic studies on D-amino acid oxidase x substrate complex: implications of the crystal structure of enzyme x substrate analog complex.
J.Biochem.(Tokyo), 122, 1997
|
|
4IJ5
 
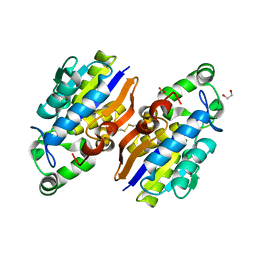 | | Crystal Structure of a Novel-type Phosphoserine Phosphatase from Hydrogenobacter thermophilus TK-6 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Phosphoserine phosphatase 1 | | Authors: | Chiba, Y, Horita, S, Ohtsuka, J, Arai, H, Nagata, K, Igarashi, Y, Tanokura, M, Ishii, M. | | Deposit date: | 2012-12-21 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural units important for activity of a novel-type phosphoserine phosphatase from Hydrogenobacter thermophilus TK-6 revealed by crystal structure analysis
J.Biol.Chem., 288, 2013
|
|
1IY6
 
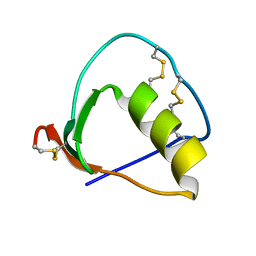 | | Solution structure of OMSVP3 variant, P14C/N39C | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1IY5
 
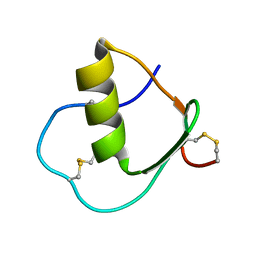 | | Solution structure of wild type OMSVP3 | | Descriptor: | OMSVP3 | | Authors: | Hemmi, H, Kumazaki, T, Yamazaki, T, Kojima, S, Yoshida, T, Kyogoku, Y, Katsu, M, Yokosawa, H, Miura, K, Kobayashi, Y. | | Deposit date: | 2002-07-23 | | Release date: | 2003-03-11 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Inhibitory Specificity Change of Ovomucoid Third Domain of the Silver Pheasant upon Introduction of an Engineered Cys14-Cys39 Bond
BIOCHEMISTRY, 42, 2003
|
|
1CMJ
 
 | |
6AKG
 
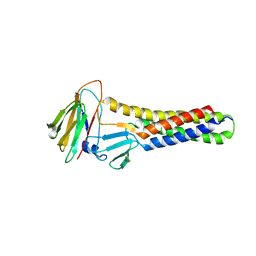 | |
1I2H
 
 | | CRYSTAL STRUCTURE ANALYSIS OF PSD-ZIP45(HOMER1C/VESL-1L)CONSERVED HOMER 1 DOMAIN | | Descriptor: | PSD-ZIP45(HOMER-1C/VESL-1L) | | Authors: | Irie, K, Nakatsu, T, Mitsuoka, K, Fujiyoshi, Y, Kato, H. | | Deposit date: | 2001-02-09 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Homer 1 Family Conserved Region Reveals the Interaction Between the EVH1 Domain and
Own Proline-rich Motif
J.Mol.Biol., 318, 2002
|
|
6AKE
 
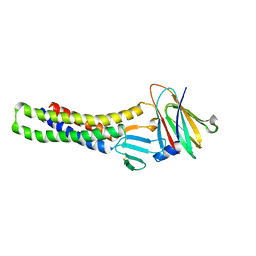 | |
1IQP
 
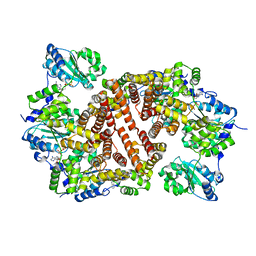 | | Crystal Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RFCS | | Authors: | Oyama, T, Ishino, Y, Cann, I.K.O, Ishino, S, Morikawa, K. | | Deposit date: | 2001-07-24 | | Release date: | 2001-09-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic Structure of the Clamp Loader Small Subunit from Pyrococcus furiosus
Mol.Cell, 8, 2001
|
|
3WFE
 
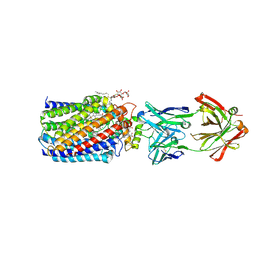 | | Reduced and cyanide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CYANIDE ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
8JLB
 
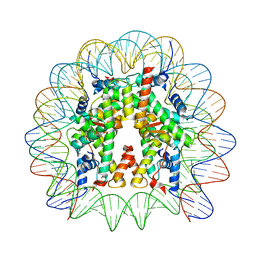 | | Cryo-EM structure of the 145 bp human nucleosome containing H3.2 C110A mutant | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JL9
 
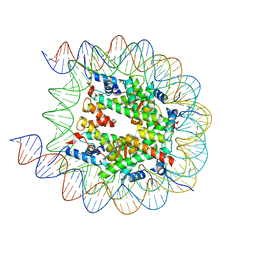 | | Cryo-EM structure of the human nucleosome with scFv | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLD
 
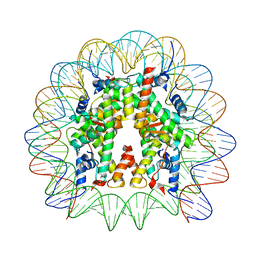 | | Cryo-EM structure of the 145 bp human nucleosome containing acetylated H3 tail | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
8JLA
 
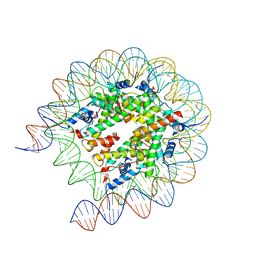 | | Cryo-EM structure of the human nucleosome lacking N-terminal region of H2A, H2B, H3, and H4 | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Oishi, T, Hatazawa, S, Kujirai, T, Kato, J, Kobayashi, Y, Ogasawara, M, Akatsu, M, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Contributions of histone tail clipping and acetylation in nucleosome transcription by RNA polymerase II.
Nucleic Acids Res., 51, 2023
|
|
3WFC
 
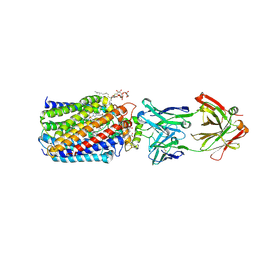 | | Reduced and carbonmonoxide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
3WFD
 
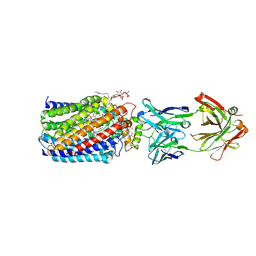 | | Reduced and acetaldoxime-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | (1E)-N-hydroxyethanimine, CALCIUM ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
1UFZ
 
 | | Solution structure of HBS1-like domain in hypothetical protein BAB28515 | | Descriptor: | Hypothetical protein BAB28515 | | Authors: | He, F, Muto, Y, Hayami, N, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HBS1-like domain in hypothetical protein BAB28515
To be Published
|
|
