1ZPB
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with 4-Methyl-pentanoic acid {1-[4-guanidino-1-(thiazole-2-carbonyl)-butylcarbamoyl]-2-methyl-propyl}-amide | | Descriptor: | 4-METHYL-PENTANOIC ACID {1-[4-GUANIDINO-1-(THIAZOLE-2-CARBONYL)-BUTYLCARBAMOYL]-2-METHYL-PROPYL}-AMIDE, Coagulation factor XI, SULFATE ION | | Authors: | Deng, H, Bannister, T.D, Jin, L, Nagafuji, P, Celatka, C.A, Lin, J, Lazarova, T.I, Rynkiewicz, M.J, Quinn, J, Bibbins, F, Pandey, P, Gorga, J, Babine, R.E, Meyers, H.V, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-05-16 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis, SAR exploration, and X-ray crystal structures of factor XIa inhibitors containing an alpha-ketothiazole arginine
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1T3H
 
 | | X-ray Structure of Dephospho-CoA Kinase from E. coli Norteast Structural Genomics Consortium Target ER57 | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | Kuzin, A.P, Chen, Y, Forouhar, F, Edstrom, W, Benach, J, Vorobiev, S, Acton, T, Shastry, R, Ma, L.-C, Xia, R, Montelione, G, Tong, L, Hunt, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-04-26 | | Release date: | 2004-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray Structure of Dephospho-CoA Kinase from E. coli
Norteast Structural Genomics Consortium Target ER57
TO BE PUBLISHED
|
|
5EJL
 
 | | MrkH, A novel c-di-GMP dependence transcription regulatory factor. | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Klebsiella pneumoniae genome assembly NOVST, ... | | Authors: | Wang, F, Zhu, D, Gu, L. | | Deposit date: | 2015-11-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The PilZ domain of MrkH represents a novel DNA binding motif
Protein Cell, 7, 2016
|
|
1YVK
 
 | | Crystal Structure of the Bacillis subtilis Acetyltransferase in complex with CoA, Northeast Structural Genomics Target SR237. | | Descriptor: | COENZYME A, hypothetical protein BSU33890 | | Authors: | Forouhar, F, Yong, W, Xiao, R, Ciano, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-02-15 | | Release date: | 2005-02-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of the Bacillis subtilis Acetyltransferase in complex with CoA, Northeast Structural Genomics Target SR237.
To be Published
|
|
1TDK
 
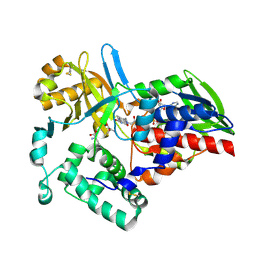 | |
6D0P
 
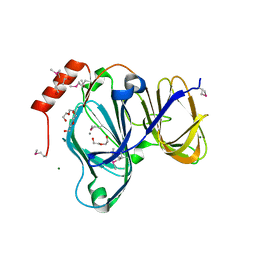 | | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Dubrovska, I, Kiryukhina, O, Endres, M, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Quercetin 2,3-dioxygenase from Acinetobacter baumannii
To be Published
|
|
8Q2H
 
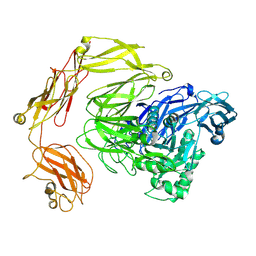 | | beta-galactosidase from Bacillus circulans | | Descriptor: | beta-galactosidase | | Authors: | Kascakova, B, Hovorkova, M, Petraskova, L, Novacek, J, Pinkas, D, Gardian, Z, Kren, V, Bojarova, P, Kuta Smatanova, I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The variable structural flexibility of the Bacillus circulans beta-galactosidase isoforms determines their unique functionalities.
Structure, 2024
|
|
1TEM
 
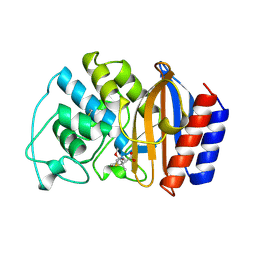 | | 6 ALPHA HYDROXYMETHYL PENICILLOIC ACID ACYLATED ON THE TEM-1 BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | 2-(1-CARBOXY-2-HYDROXY-ETHYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, TEM-1 BETA LACTAMASE | | Authors: | Maveyraud, L, Massova, I, Samama, J.P, Mobashery, S. | | Deposit date: | 1996-05-28 | | Release date: | 1997-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 6Alpha-Hydroxymethylpenicillanate Complexed to the Tem-1 Beta-Lactamase from Escherichia Coli: Evidence on the Mechanism of Action of a Novel Inhibitor Designed by a Computer-Aided Process
J.Am.Chem.Soc., 118, 1996
|
|
5EPA
 
 | | Crystal structure of non-heme alpha ketoglutarate dependent carbocyclase SnoK from nogalamycin biosynthesis | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Selvaraj, B, Lindqvist, Y, Siitonen, V, Niiranen, L, Metsa-Ketela, M, Schneider, G. | | Deposit date: | 2015-11-11 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Divergent non-heme iron enzymes in the nogalamycin biosynthetic pathway.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6CMH
 
 | | SYNTHETIC LINEAR MODIFIED ENDOTHELIN-1 AGONIST | | Descriptor: | PROTEIN (ENDOTHELIN-1) | | Authors: | Hewage, C.M, Jiang, L, Parkinson, J.A, Ramage, R, Sadler, I.H. | | Deposit date: | 1998-09-03 | | Release date: | 1999-09-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a novel ETB receptor selective agonist ET1-21 [Cys(Acm)1,15, Aib3,11, Leu7] by nuclear magnetic resonance spectroscopy and molecular modelling.
J.Pept.Res., 53, 1999
|
|
1TG9
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-28 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
8QJ7
 
 | |
8QFC
 
 | | UFL1 E3 ligase bound 60S ribosome | | Descriptor: | 60S ribosomal protein L10a, CDK5 regulatory subunit-associated protein 3, DDRGK domain-containing protein 1, ... | | Authors: | Makhlouf, L, Zeqiraj, E, Kulathu, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Nature, 627, 2024
|
|
5EHY
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 4-(furan-3-yl)-3-phenyl-2~{H}-pyrazolo[4,3-c]pyridine, ... | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
5EI8
 
 | | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle kinase 1 (MPS1) Using a Structure-Based Hydridization Approach | | Descriptor: | DIMETHYL SULFOXIDE, Dual specificity protein kinase TTK, ~{N}-[2-methoxy-4-(1-methylpyrazol-4-yl)phenyl]-8-(1-methylpyrazol-4-yl)pyrido[3,4-d]pyrimidin-2-amine | | Authors: | Innocenti, P, Woodward, H.L, Solanki, S, Naud, N, Westwood, I.M, Cronin, N, Hayes, A, Roberts, J, Henley, A.T, Baker, R, Faisal, A, Mak, G, Box, G, Valenti, M, De Haven Brandon, A, O'Fee, L, Saville, J, Schmitt, J, Burke, R, van Montfort, R.L.M, Raymaud, F.I, Eccles, S.A, Linardopoulos, S, Blagg, J, Hoelder, S. | | Deposit date: | 2015-10-29 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Rapid Discovery of Pyrido[3,4-d]pyrimidine Inhibitors of Monopolar Spindle Kinase 1 (MPS1) Using a Structure-Based Hybridization Approach.
J.Med.Chem., 59, 2016
|
|
1TKQ
 
 | | SOLUTION STRUCTURE OF A LINKED UNSYMMETRIC GRAMICIDIN IN A MEMBRANE-ISOELECTRICAL SOLVENTS MIXTURE IN THE PRESENCE OF CsCl | | Descriptor: | GRAMICIDIN A, MINI-GRAMICIDIN A, SUCCINIC ACID | | Authors: | Xie, X, Al-Momani, L, Bockelmann, D, Griesinger, C, Koert, U. | | Deposit date: | 2004-06-09 | | Release date: | 2004-07-13 | | Last modified: | 2019-09-25 | | Method: | SOLUTION NMR | | Cite: | An Asymmetric Ion Channel Derived from Gramicidin A. Synthesis, Function and NMR Structure.
FEBS J., 272, 2005
|
|
1T7W
 
 | | Zn-alpha-2-glycoprotein; CHO-ZAG PEG 400 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Zinc-alpha-2-glycoprotein | | Authors: | Delker, S.L, West Jr, A.P, McDermott, L, Kennedy, M.W, Bjorkman, P.J. | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of ligand binding by Zn-alpha2-glycoprotein.
J.Struct.Biol., 148, 2004
|
|
5EIV
 
 | | Crystal structure of complex of osteoclast-associated immunoglobulin-like receptor (OSCAR) and a synthetic collagen consensus peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhou, L, Blaszczyk, M, Chirgadze, D, Bihan, D, Farndale, R.W. | | Deposit date: | 2015-10-30 | | Release date: | 2015-11-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | Structural basis for collagen recognition by the immune receptor OSCAR.
Blood, 127, 2016
|
|
1Z5R
 
 | | Crystal Structure of Activated Porcine Pancreatic Carboxypeptidase B | | Descriptor: | ZINC ION, procarboxypeptidase B | | Authors: | Adler, M, Bryant, J, Buckman, B, Islam, I, Larsen, B, Finster, S, Kent, L, May, K, Mohan, R, Yuan, S, Whitlow, M. | | Deposit date: | 2005-03-18 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structures of potent thiol-based inhibitors bound to carboxypeptidase b.
Biochemistry, 44, 2005
|
|
1Z2Z
 
 | | Crystal Structure of the Putative tRNA pseudouridine synthase D (TruD) from Methanosarcina mazei, Northeast Structural Genomics Target MaR1 | | Descriptor: | Probable tRNA pseudouridine synthase D | | Authors: | Forouhar, F, Yong, W, Ciano, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-03-10 | | Release date: | 2005-04-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Putative tRNA pseudouridine synthase D (TruD) from Methanosarcina mazei, Northeast Structural Genomics Target MaR1
To be Published
|
|
4GBQ
 
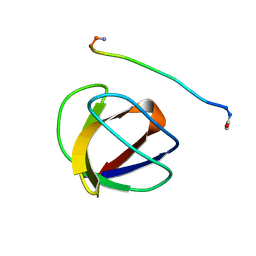 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1TBT
 
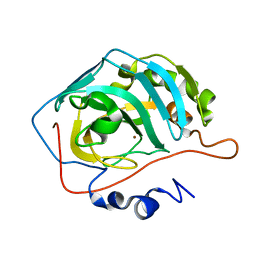 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-20 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II.
Biochemistry, 44, 2005
|
|
6DW4
 
 | | SAMHD1 Bound to Cladribine-TP in the Catalytic Pocket and Allosteric Pocket | | Descriptor: | 2'-deoxy-2-methyladenosine 5'-(tetrahydrogen triphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1SVE
 
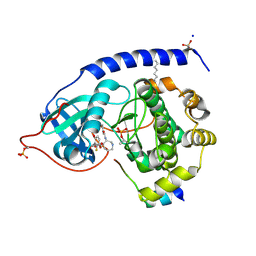 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 1 | | Descriptor: | (4R)-4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZOIC ACID (3R)-3-[(PYRIDINE-4-CARBONYL)AMINO]-AZEPAN-4-YL ESTER, N-OCTANOYL-N-METHYLGLUCAMINE, SODIUM ION, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
1Z6B
 
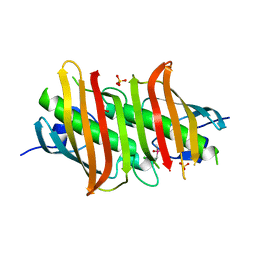 | | Crystal structure of Plasmodium falciparum FabZ at 2.1 A | | Descriptor: | CACODYLATE ION, CHLORIDE ION, SULFATE ION, ... | | Authors: | Kostrewa, D, Winkler, F.K, Folkers, G, Scapozza, L, Perozzo, R. | | Deposit date: | 2005-03-22 | | Release date: | 2005-06-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The crystal structure of PfFabZ, the unique beta-hydroxyacyl-ACP dehydratase involved in fatty acid biosynthesis of Plasmodium falciparum
PROTEIN SCI., 14, 2005
|
|
