6LAH
 
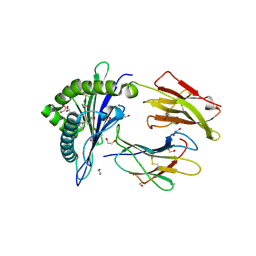 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with lysophosphatidylcholine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
8RS5
 
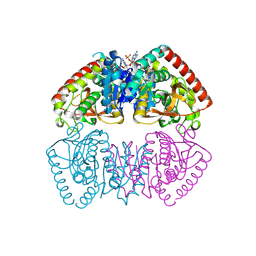 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Coquille, S, Engilberge, S, Madern, D. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Allostery and Evolution: A Molecular Journey Through the Structural and Dynamical Landscape of an Enzyme Super Family.
Mol.Biol.Evol., 42, 2025
|
|
8RWL
 
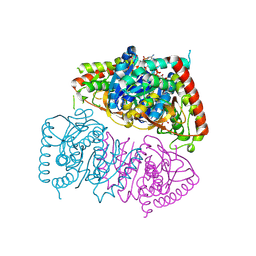 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Malate dehydrogenase, ... | | Authors: | Coquille, S, Roche, J, Engilberge, S, Girard, E, Madern, D. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allostery and Evolution: A Molecular Journey Through the Structural and Dynamical Landscape of an Enzyme Super Family.
Mol.Biol.Evol., 42, 2025
|
|
6LH9
 
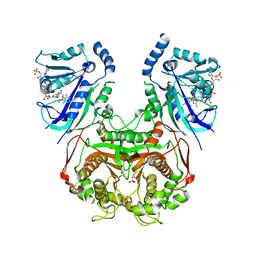 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 46 and NADPH | | Descriptor: | 2-[[4,6-bis(azanyl)-2,2-dimethyl-1,3,5-triazin-1-yl]oxy]-N-(4-chlorophenyl)ethanamide, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-12-07 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
6LHX
 
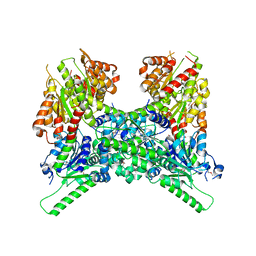 | | Crystal structure of ThsA | | Descriptor: | ThsA | | Authors: | Bae, E, Ka, D, Oh, H. | | Deposit date: | 2019-12-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and functional evidence of bacterial antiphage protection by Thoeris defense system via NAD+degradation.
Nat Commun, 11, 2020
|
|
8RMJ
 
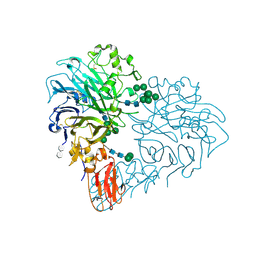 | | Drosophila Semaphorin 2b in complex with glycosaminoglycan mimic SOS | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nourisanami, F, Sobol, M, Rozbesky, D. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Molecular mechanisms of proteoglycan-mediated semaphorin signaling in axon guidance.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8RG0
 
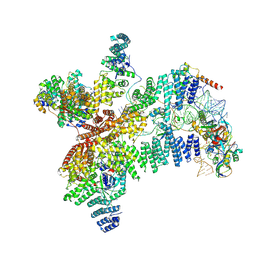 | | Structure of human eIF3 core from closed 48S translation initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S13, 40S ribosomal protein S14, ... | | Authors: | Petrychenko, V, Yi, S.-H, Liedtke, D, Peng, B.Z, Rodnina, M.V, Fischer, N. | | Deposit date: | 2023-12-13 | | Release date: | 2024-08-14 | | Last modified: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for translational control by the human 48S initiation complex.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6L2O
 
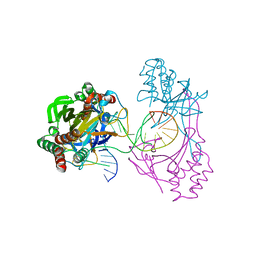 | | Crystal structure of the R.PabI(Y68F-K154A)-dsDNA(GTAC-5bp-GTAC) complex | | Descriptor: | DNA (5'-D(*CP*A*GP*CP*AP*GP*TP*AP*CP*TP*TP*AP*AP*AP*GP*TP*AP*CP*TP*GP*CP*TP*G)-3'), RE_R_Pab1 domain-containing protein | | Authors: | Miyazono, K, Wang, D, Ito, T, Tanokura, M. | | Deposit date: | 2019-10-05 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distortion of double-stranded DNA structure by the binding of the restriction DNA glycosylase R.PabI.
Nucleic Acids Res., 48, 2020
|
|
6LAM
 
 | | Crystal structure of rhesus macaque MHC class I molecule Mamu-B*098 complexed with lysophosphatidylethanolamine | | Descriptor: | (2R)-2,3-dihydroxypropyl hexadecanoate, 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Shima, Y, Morita, D. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of lysophospholipid-bound MHC class I molecules.
J.Biol.Chem., 295, 2020
|
|
6LB4
 
 | | Crystal structure of dimeric RXR-LBD complexed with NEt-3ME and TIF2 co-activator | | Descriptor: | 1,2-ETHANEDIOL, 6-[ethyl-[3-(2-methoxyethoxy)-4-propan-2-yl-phenyl]amino]pyridine-3-carboxylic acid, Nuclear receptor coactivator 2, ... | | Authors: | Imai, D, Numoto, N, Nakano, S, Kakuta, H, Ito, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of dimeric RXR-LBD complexed with NEt-3ME and TIF2 co-activator
To Be Published
|
|
6L84
 
 | | Complex of DNA polymerase IV and D-DNA duplex | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), DNA (5'-D(P*CP*GP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Chung, H.S, An, J, Hwang, D. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The crystal structure of a natural DNA polymerase complexed with mirror DNA.
Chem.Commun.(Camb.), 56, 2020
|
|
8S5R
 
 | | Structure of the Chlamydia pneumoniae effector SemD | | Descriptor: | Effector SemD | | Authors: | Kocher, F, Applegate, V, Reiners, J, Port, A, Spona, D, Haensch, S, Smits, S.H, Hegemann, J, Moelleken, K. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Chlamydia pneumoniae effector SemD exploits its host's endocytic machinery by structural and functional mimicry.
Nat Commun, 15, 2024
|
|
8S1M
 
 | | Crystal structure of human GABARAP fused to EGFR (1076-1099) | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor,Gamma-aminobutyric acid receptor-associated protein, GLYCEROL | | Authors: | Ueffing, A, Willbold, D, Weiergraeber, O.H. | | Deposit date: | 2024-02-15 | | Release date: | 2024-08-14 | | Last modified: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GABARAP interacts with EGFR - supporting the unique role of this hAtg8 protein during receptor trafficking.
Febs Lett., 598, 2024
|
|
8S5T
 
 | | Structure of SemD in complex | | Descriptor: | Effector SemD, Neural Wiskott-Aldrich syndrome protein | | Authors: | Kocher, F, Applegate, V, Port, A, Reiners, J, Spona, D, Haensch, S, Smits, S.H, Hegemann, J, Moelleken, K. | | Deposit date: | 2024-02-25 | | Release date: | 2024-08-07 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Chlamydia pneumoniae effector SemD exploits its host's endocytic machinery by structural and functional mimicry.
Nat Commun, 15, 2024
|
|
6LB6
 
 | | Crystal structure of dimeric RXR-LBD complexed with partial agonist NEt-4IB and TIF2 co-activator | | Descriptor: | 6-[ethyl-[4-(2-methylpropoxy)-3-propan-2-yl-phenyl]amino]pyridine-3-carboxylic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Imai, D, Numoto, N, Nakano, S, Kakuta, H, Ito, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the full and partial agonist activities of retinoid X receptor alpha ligands with an iso-butoxy and an isopropyl group
Biochem.Biophys.Res.Commun., 734, 2024
|
|
8S33
 
 | | Malic semialdehyde dehydrogenase (MSA-DH) from Acinetobacter baumannii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Succinate-semialdehyde dehydrogenase [NAD(P)+] | | Authors: | Piskol, F, Lukat, P, Blankenfeldt, W, Jahn, D, Moser, J. | | Deposit date: | 2024-02-19 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Biochemical and structural elucidation of the L-carnitine degradation pathway of the human pathogen Acinetobacter baumannii .
Front Microbiol, 15, 2024
|
|
6YT9
 
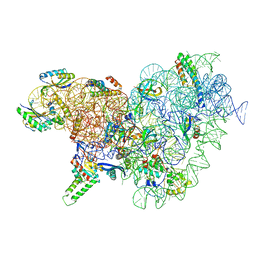 | | Acinetobacter baumannii ribosome-tigecycline complex - 30S subunit body | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Nicholson, D, Edwards, T.A, O'Neill, A.J, Ranson, N.A. | | Deposit date: | 2020-04-24 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of the 70S Ribosome from the Human Pathogen Acinetobacter baumannii in Complex with Clinically Relevant Antibiotics.
Structure, 28, 2020
|
|
6LE9
 
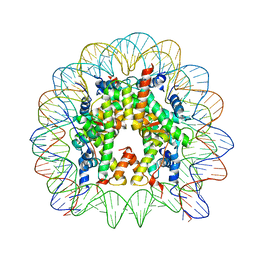 | | The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-K, Histone H3.1, ... | | Authors: | Soman, A, Liew, C.W, Teo, H.L, Berezhnoy, N, Korolev, N, Rhodes, D, Nordenskiold, L. | | Deposit date: | 2019-11-24 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The human telomeric nucleosome displays distinct structural and dynamic properties.
Nucleic Acids Res., 48, 2020
|
|
6LDM
 
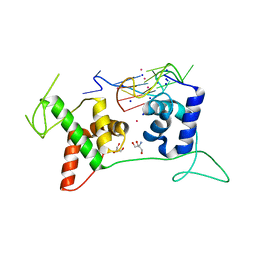 | | Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA-binding protein RAP1, G-guadruplex DNA, ... | | Authors: | Traczyk, A, Gill, D.J, Chong, W.L, Rhodes, D. | | Deposit date: | 2019-11-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1.
Nucleic Acids Res., 48, 2020
|
|
6LEZ
 
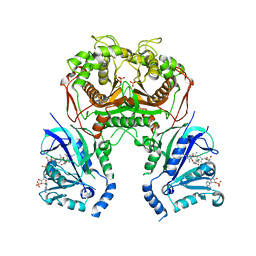 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with compound 46 and NADPH | | Descriptor: | 2-[[4,6-bis(azanyl)-2,2-dimethyl-1,3,5-triazin-1-yl]oxy]-N-(4-chlorophenyl)ethanamide, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-11-27 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.644 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
6LHI
 
 | | Quadruple mutant (N51I+C59R+S108N+I164L) plasmodium falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS) complexed with C466 (compound 42) and NADPH | | Descriptor: | 1-[3-[(4-chlorophenyl)-(phenylmethyl)amino]propoxy]-6,6-dimethyl-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Vitsupakorn, D. | | Deposit date: | 2019-12-09 | | Release date: | 2020-12-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Flexible diaminodihydrotriazine inhibitors of Plasmodium falciparum dihydrofolate reductase: Binding strengths, modes of binding and their antimalarial activities.
Eur.J.Med.Chem., 195, 2020
|
|
7O8F
 
 | | NmHR dark state structure determined by serial femtosecond crystallography | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
7O8H
 
 | | NmHR light state structure at 10 ns after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
6LIM
 
 | | BRD4-BD1 bound with compound 40 | | Descriptor: | (3~{R})-6-[(4-isoquinolin-4-ylpyrimidin-2-yl)amino]-1,3-dimethyl-4-propan-2-yl-3~{H}-quinoxalin-2-one, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D, Li, Y. | | Deposit date: | 2019-12-12 | | Release date: | 2020-12-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75909507 Å) | | Cite: | BRD4-BD1 bound with compound 40
To Be Published
|
|
7O8G
 
 | | NmHR light state structure at 10 ps after photoexcitation determined by serial femtosecond crystallography (with extrapolated, dark and light dataset) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHLORIDE ION, Chloride pumping rhodopsin, ... | | Authors: | Mous, S, Gotthard, G, Ehrenberg, D, Sen, S, James, D, Johnson, P, Weinert, T, Nass, K, Furrer, A, Kekilli, D, Ma, P, Bruenle, S, Casadei, C, Martiel, I, Dworkowski, F, Gashi, D, Skopintsev, P, Wranik, M, Knopp, G, Panepucci, E, Panneels, V, Cirelli, C, Ozerov, D, Schertler, G, Wang, M, Milne, C, Standfuss, J, Schapiro, I, Heberle, J, Nogly, P. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Dynamics and mechanism of a light-driven chloride pump.
Science, 375, 2022
|
|
