7LPN
 
 | |
8Y42
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) in complex with compound 51 | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(1~{S},2~{R})-2-[[4-cyclopropyl-2-(methylcarbamoyl)-6-nitro-phenyl]amino]cyclohexyl]-2-oxidanylidene-1~{H}-quinoline-4-carboxamide | | Authors: | Nie, T.Q, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-01-30 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Novel Nonpeptidic and Noncovalent Small Molecule 3CL pro Inhibitors as anti-SARS-CoV-2 Drug Candidate.
J.Med.Chem., 67, 2024
|
|
8Y44
 
 | | Crystal structure of SARS-CoV-2 3CL protease (3CLpro) in complex with compound 44 | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(1~{S},2~{R})-2-[(4-bromanyl-2-morpholin-4-ylcarbonyl-6-nitro-phenyl)amino]cyclohexyl]-2-oxidanylidene-1~{H}-quinoline-4-carboxamide | | Authors: | Nie, T.Q, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2024-01-30 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of Novel Nonpeptidic and Noncovalent Small Molecule 3CL pro Inhibitors as anti-SARS-CoV-2 Drug Candidate.
J.Med.Chem., 67, 2024
|
|
8XEW
 
 | | Cryo-EM structure of defence-associatedsirtuin 2 (DSR2) H171A protein | | Descriptor: | DSR2 H171A | | Authors: | Li, Y, Zhang, H, Zheng, Q, Wu, Y, Li, S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Structural insights into activation mechanisms on NADase of the bacterial DSR2 anti-phage defense system.
Sci Adv, 10, 2024
|
|
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
1ZBN
 
 | |
3GMJ
 
 | | Crystal structure of MAD MH2 domain | | Descriptor: | Protein mothers against dpp | | Authors: | Wu, J.W, Wang, C. | | Deposit date: | 2009-03-14 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the MH2 domain of Drosophila Mad
SCI.CHINA, SER.C: LIFE SCI., 52, 2009
|
|
6IE2
 
 | | Crystal structure of methyladenine demethylase | | Descriptor: | 2-OXOGLUTARIC ACID, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Tian, L.F, Tang, Q, Chen, Z.Z, Yan, X.X. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1.
Cell Res., 30, 2020
|
|
8WWC
 
 | | De novo design binder of HRAS -120-4 | | Descriptor: | De novo design protein 120-4, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Wang, L, Wang, S, Liu, Y. | | Deposit date: | 2023-10-25 | | Release date: | 2024-10-09 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | De novo protein design with a denoising diffusion network independent of pretrained structure prediction models.
Nat.Methods, 21, 2024
|
|
3HOB
 
 | |
2EB3
 
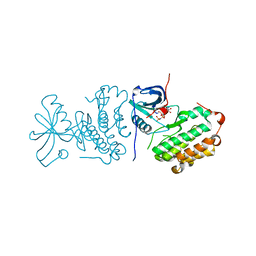 | | Crystal structure of mutated EGFR kinase domain (L858R) in complex with AMPPNP | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Senba, K, Yamamoto, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 2012
|
|
3IKA
 
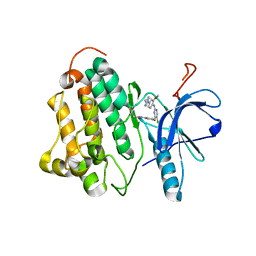 | |
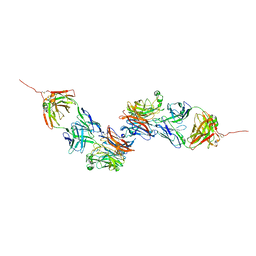
8K0D
 
 | |
8K0C
 
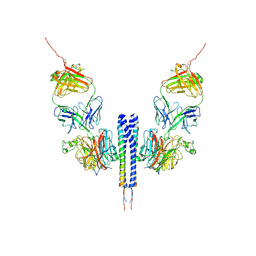 | |
1ETG
 
 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, 19 STRUCTURES | | Descriptor: | REV PEPTIDE, REV RESPONSIVE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
1ETF
 
 | | REV RESPONSE ELEMENT (RRE) RNA COMPLEXED WITH REV PEPTIDE, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REV PEPTIDE, REV RESPONSE ELEMENT RNA | | Authors: | Battiste, J.L, Mao, H, Rao, N.S, Tan, R, Muhandiram, D.R, Kay, L.E, Frankel, A.D, Willamson, J.R. | | Deposit date: | 1996-08-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex.
Science, 273, 1996
|
|
5ZHE
 
 | | STRUCTURE OF E. COLI UNDECAPRENYL DIPHOSPHATE SYNTHASE IN COMPLEX WITH BPH-981 | | Descriptor: | 2-hydroxy-6-(tetradecyloxy)benzoic acid, Ditrans,polycis-undecaprenyl-diphosphate synthase ((2E,6E)-farnesyl-diphosphate specific) | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Ko, T.P, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-03-13 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Lipophilic Bisphosphonates That Target Bacterial Cell Wall and Quinone Biosynthesis.
J.Med.Chem., 62, 2019
|
|
6IE3
 
 | | Crystal structure of methyladenine demethylase | | Descriptor: | ETHANOL, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | Authors: | Tian, L.F, Tang, Q, Chen, Z.Z, Yan, X.X. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of nucleic acid recognition and 6mA demethylation by human ALKBH1.
Cell Res., 30, 2020
|
|
7XOU
 
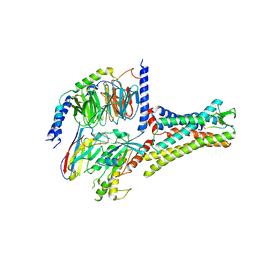 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | CCK-8, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOV
 
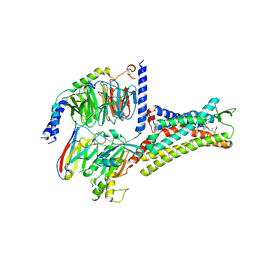 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | 2-[2-[[4-(4-chloranyl-2,5-dimethoxy-phenyl)-5-(2-cyclohexylethyl)-1,3-thiazol-2-yl]carbamoyl]-5,7-dimethyl-indol-1-yl]ethanoic acid, Cholecystokinin receptor type A, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
7XOW
 
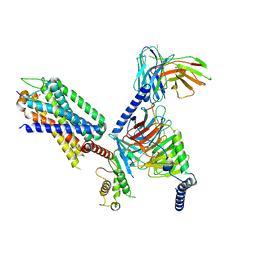 | | Structural insights into human brain gut peptide cholecystokinin receptors | | Descriptor: | Gastrin, Gastrin/cholecystokinin type B receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ding, Y, Zhang, H, Liao, Y, Chen, L, Ji, S. | | Deposit date: | 2022-05-01 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into human brain-gut peptide cholecystokinin receptors.
Cell Discov, 8, 2022
|
|
3HNB
 
 | |
1AAX
 
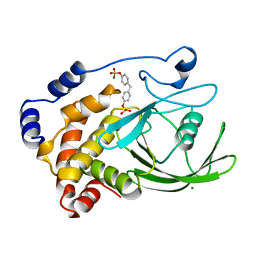 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO BIS(PARA-PHOSPHOPHENYL)METHANE (BPPM) MOLECULES | | Descriptor: | 4-PHOSPHONOOXY-PHENYL-METHYL-[4-PHOSPHONOOXY]BENZEN, MAGNESIUM ION, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Puius, Y.A, Zhao, Y, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | Deposit date: | 1997-01-16 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
7DRI
 
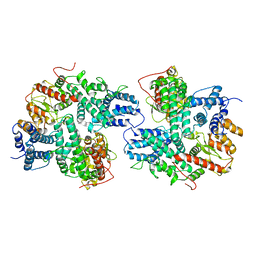 | | Structure of SspE_CTD_41658 | | Descriptor: | DUF1524 domain | | Authors: | Haiyan, G, Jinchuan, Z, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2020-12-28 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Nicking mechanism underlying the DNA phosphorothioate-sensing antiphage defense by SspE.
Nat Commun, 13, 2022
|
|
1ARJ
 
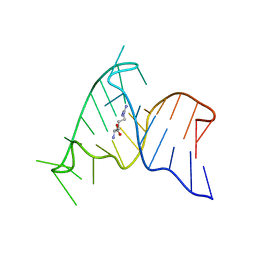 | | ARG-BOUND TAR RNA, NMR | | Descriptor: | ARGININE, TAR RNA | | Authors: | Aboul-Ela, F, Varani, G, Karn, J. | | Deposit date: | 1995-08-30 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein.
J.Mol.Biol., 253, 1995
|
|
