3HCW
 
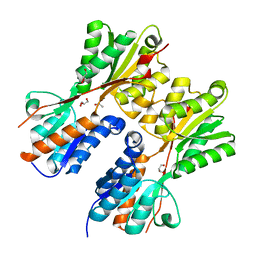 | | CRYSTAL STRUCTURE OF PROBABLE maltose operon transcriptional repressor malR FROM STAPHYLOCOCCUS AREUS | | Descriptor: | GLYCEROL, Maltose operon transcriptional repressor | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Maltose Operon Transcriptional Repressor from Staphylococcus Aureus
To be Published
|
|
4ESK
 
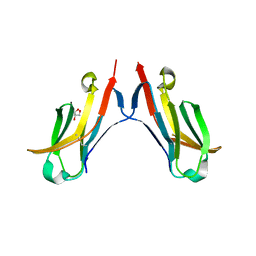 | |
4E6P
 
 | |
3HIQ
 
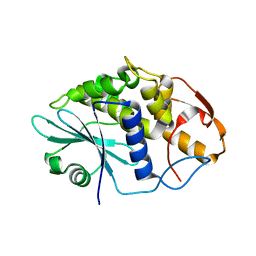 | |
4ECJ
 
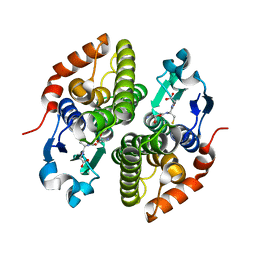 | | Crystal structure of glutathione s-transferase prk13972 (target efi-501853) from pseudomonas aeruginosa pacs2 complexed with glutathione | | Descriptor: | GLUTATHIONE, glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Prk13972 from Pseudomonas Aeruginosa
To be Published
|
|
4EXJ
 
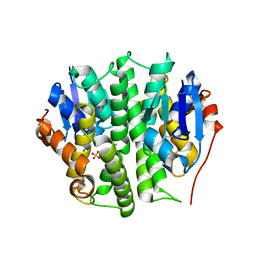 | | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of glutathione s-transferase like protein lelg_03239 (target efi-501752) from lodderomyces elongisporus
To be Published
|
|
3HIT
 
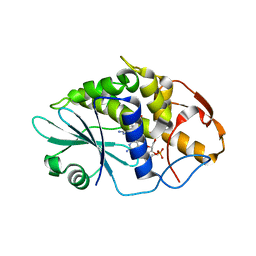 | | Crystal structure of Saporin-L1 in complex with the dinucleotide inhibitor, a transition state analogue | | Descriptor: | 5'-O-[(S)-{[(3R,4R)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[(S)-hydroxy(3-hydroxypropoxy)phosphoryl]oxy}methyl)pyrrolidin-3-yl]oxy}(hydroxy)phosphoryl]-3'-O-[(R)-hydroxy(4-hydroxybutoxy)phosphoryl]-2'-O-methylguanosine, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HG7
 
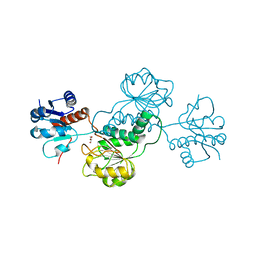 | | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449 | | Descriptor: | D-isomer specific 2-hydroxyacid dehydrogenase family protein, GLYCEROL | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF D-isomer specific 2-hydroxyacid dehydrogenase family protein from Aeromonas salmonicida subsp. salmonicida A449
To be Published
|
|
3HJG
 
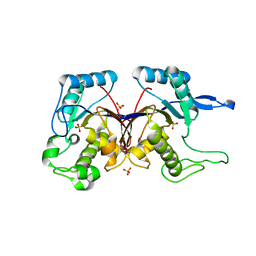 | | Crystal structure of putative alpha-ribazole-5'-phosphate phosphatase CobC FROM Vibrio parahaemolyticus | | Descriptor: | Putative alpha-ribazole-5'-phosphate phosphatase CobC, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Morano, C, Rutter, M, Iizuka, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of putative alpha-ribazole-5' -phosphate phosphatase FROM Vibrio parahaemolyticus
To be Published
|
|
3HIO
 
 | | Crystal structure of Ricin A-chain in complex with the cyclic tetranucleotide inhibitor, a transition state analogue | | Descriptor: | 9,9'-{(2R,3R,3aR,5S,7aR,9R,10R,10aR,12S,23R,25aR,27R,28R,28aR,30S,32aR,35aR,37S,39aR)-9-(6-amino-9H-purin-9-yl)-34-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-5,12,23,30,37-pentahydroxy-3,10,28-trimethoxy-5,12,23,30,37-pentaoxidotetracosahydro-2H,7H,25H-trifuro[3,2-f:3',2'-l:3'',2''-x]pyrrolo[3,4-r][1,3,5,9,11,15,17,21,23,27,29,2,4,10,16,22,28]undecaoxazapentaphosphacyclopentatriacontine-2,27-diyl}bis(2-amino-3,9-dihydro-6H-purin-6-one), Ricin, SULFATE ION | | Authors: | Ho, M, Sturm, M.B, Goldman, J.D, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIV
 
 | | Crystal structure of Saporin-L1 in complex with the trinucleotide inhibitor, a transition state analogue | | Descriptor: | (2R,3R,4R,5R)-5-(2-amino-6-oxo-3,6-dihydro-9H-purin-9-yl)-2-({[(S)-({(3R,4R)-4-({[(S)-{[(2R,3R,4R,5R)-5-(2-amino-6-oxo-6,8-dihydro-9H-purin-9-yl)-2-(hydroxymethyl)-4-methoxytetrahydrofuran-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]pyrrolidin-3-yl}oxy)(hydroxy)phosphoryl]oxy}methyl)-4-methoxytetrahydrofuran-3-yl 3-hydroxypropyl hydrogen (S)-phosphate, Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HIN
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase from Rhodopseudomonas palustris CGA009 | | Descriptor: | Putative 3-hydroxybutyryl-CoA dehydratase | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-20 | | Release date: | 2009-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase from Rhodopseudomonas palustris CGA009
To be Published
|
|
3HIW
 
 | | Crystal structure of Saporin-L1 in complex with the cyclic tetranucleotide inhibitor, a transition state analogue | | Descriptor: | 9,9'-{(2R,3R,3aR,5S,7aR,9R,10R,10aR,12S,23R,25aR,27R,28R,28aR,30S,32aR,35aR,37S,39aR)-9-(6-amino-9H-purin-9-yl)-34-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-5,12,23,30,37-pentahydroxy-3,10,28-trimethoxy-5,12,23,30,37-pentaoxidotetracosahydro-2H,7H,25H-trifuro[3,2-f:3',2'-l:3'',2''-x]pyrrolo[3,4-r][1,3,5,9,11,15,17,21,23,27,29,2,4,10,16,22,28]undecaoxazapentaphosphacyclopentatriacontine-2,27-diyl}bis(2-amino-3,9-dihydro-6H-purin-6-one), Vacuolar saporin | | Authors: | Ho, M, Sturm, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2009-05-20 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Transition state analogues in structures of ricin and saporin ribosome-inactivating proteins.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3HG9
 
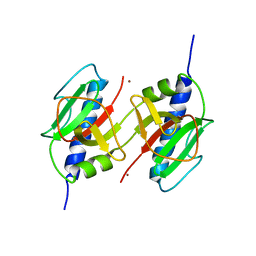 | | CRYSTAL STRUCTURE OF putative pilM protein from Pseudomonas aeruginosa 2192 | | Descriptor: | NICKEL (II) ION, PilM | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-13 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF putative pilM protein from Pseudomonas aeruginosa 2192
To be Published
|
|
4ESO
 
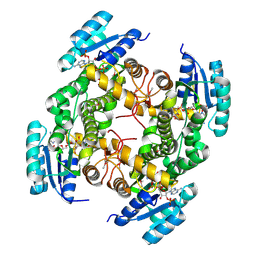 | | Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP | | Descriptor: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative oxidoreductase | | Authors: | Ghosh, A, Bhoshle, R, Toro, R, Gizzi, A, Hillerich, B, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | Crystal structure of a putative oxidoreductase protein from Sinorhizobium meliloti 1021 in complex with NADP
To be Published
|
|
4ETY
 
 | |
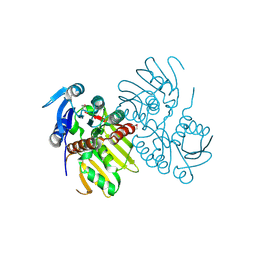
4EW6
 
 | |
3HK5
 
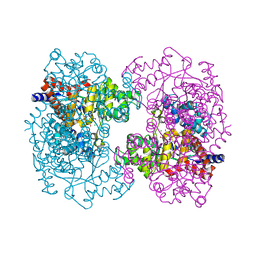 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinarate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinaric acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HK8
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Arabinohydroxamate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-arabinohydroxamic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HKA
 
 | | Crystal structure of uronate isomerase from Bacillus halodurans complexed with zinc and D-Fructuronate | | Descriptor: | CARBONATE ION, CHLORIDE ION, D-fructuronic acid, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Nguyen, T.T, Raushel, F.M, Almo, S.C. | | Deposit date: | 2009-05-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the reaction catalyzed by uronate isomerase illustrates how an isomerase may have evolved from a hydrolase within the amidohydrolase superfamily.
Biochemistry, 48, 2009
|
|
3HIS
 
 | |
3HUH
 
 | | The structure of biphenyl-2,3-diol 1,2-dioxygenase iii-related protein from salmonella typhimurium | | Descriptor: | Virulence protein STM3117 | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-14 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of biphenyl-2,3-diol 1,2-dioxygenase iii-related protein from salmonella typhimurium
To be Published
|
|
3HUR
 
 | | Crystal structure of alanine racemase from Oenococcus oeni | | Descriptor: | Alanine racemase, SULFATE ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Hu, S, Romero, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-15 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of alanine racemase from Oenococcus oeni
To be Published
|
|
4EUN
 
 | | Crystal structure of a sugar kinase (Target EFI-502144 from Janibacter sp. HTCC2649), unliganded structure | | Descriptor: | SULFATE ION, thermoresistant glucokinase | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-04-25 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a sugar kinase (Target EFI-502144 from Janibacter sp. HTCC2649), unliganded structure
To be Published
|
|
4E4F
 
 | | Crystal structure of enolase PC1_0802 (TARGET EFI-502240) from Pectobacterium carotovorum subsp. carotovorum PC1 | | Descriptor: | CHLORIDE ION, FORMIC ACID, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-12 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of ENOLASE PC1_0802 from Pectobacterium carotovorum
To be Published
|
|
