6WKX
 
 | | Cryo-EM of Form 1 related peptide filament, 15-10-3 | | Descriptor: | peptide 15-10-3 | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL0
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3-RD | | Descriptor: | peptide 36-31-3-RD | | Authors: | Wang, F, Gnewou, O.M, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6N8D
 
 | |
6WKY
 
 | | Cryo-EM of Form 1 related peptide filament, 29-24-3 | | Descriptor: | peptide 29-24-3 | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6VJJ
 
 | | Crystal Structure of wild-type KRAS4b (GMPPNP-bound) in complex with RAS-binding domain (RBD) of RAF1/CRAF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6WMI
 
 | | ZNF410 zinc fingers 1-5 with 17 mer blunt DNA Oligonucleotide | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*CP*AP*TP*AP*AP*TP*AP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*TP*AP*TP*GP*GP*GP*AP*TP*GP*TP*G)-3'), ... | | Authors: | Ren, R, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | ZNF410 Uniquely Activates the NuRD Component CHD4 to Silence Fetal Hemoglobin Expression.
Mol.Cell, 81, 2021
|
|
6WQ2
 
 | | Cryo-EM of the S. islandicus filamentous virus, SIFV | | Descriptor: | A-DNA, Structural protein MCP1, Structural protein MCP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Zheng, W, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6N81
 
 | |
6GYI
 
 | |
3ARD
 
 | | Ternary crystal structure of the mouse NKT TCR-CD1d-3'deoxy-alpha-galactosylceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
6G4Z
 
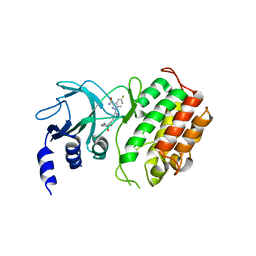 | | Crystal structure of murine NF-kappaB inducing kinase (NIK) in complex with compound 2f | | Descriptor: | 5-fluoranyl-1-[4-[2-[(3~{R})-1-methyl-3-oxidanyl-2-oxidanylidene-pyrrol-3-yl]ethynyl]pyridin-2-yl]indazole-3-carboxamide, Mitogen-activated protein kinase kinase kinase 14 | | Authors: | Leonardo-Silvestre, H, McEwan, P.A, Hymowitz, S.G. | | Deposit date: | 2018-03-28 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Scaffold-Hopping Approach To Discover Potent, Selective, and Efficacious Inhibitors of NF-kappa B Inducing Kinase.
J. Med. Chem., 61, 2018
|
|
3ARE
 
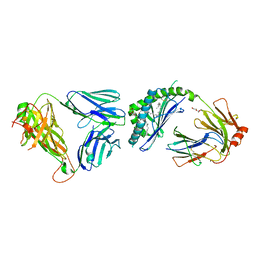 | | Ternary crystal structure of the mouse NKT TCR-CD1d-4'deoxy-alpha-galactosylceramide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARF
 
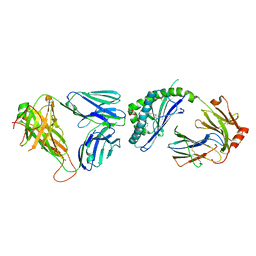 | | Ternary crystal structure of the mouse NKT TCR-CD1d-C20:2 | | Descriptor: | (11Z,14E)-N-[(2S,3S,4R)-1-(alpha-D-galactopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARG
 
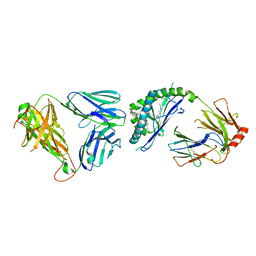 | | Ternary crystal structure of the mouse NKT TCR-CD1d-alpha-glucosylceramide(C20:2) | | Descriptor: | (11E,14E)-N-[(2S,3S,4R)-1-(alpha-D-glucopyranosyloxy)-3,4-dihydroxyoctadecan-2-yl]icosa-11,14-dienamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-27 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
3ARB
 
 | | Ternary crystal structure of the NKT TCR-CD1d-alpha-galactosylceramide analogue-OCH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wun, K.S, Rossjohn, J. | | Deposit date: | 2010-11-26 | | Release date: | 2011-03-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells
Immunity, 34, 2011
|
|
6O1F
 
 | |
6GH7
 
 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-[(3~{R})-pyrrolidin-3-yl]pentanamide, Streptavidin | | Authors: | Nodling, A.R, Tsai, Y.H, Luk, L.Y.P, Rizkallah, P, Jin, Y. | | Deposit date: | 2018-05-04 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Reactivity and Selectivity of Iminium Organocatalysis Improved by a Protein Host.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
6OFS
 
 | |
6OBQ
 
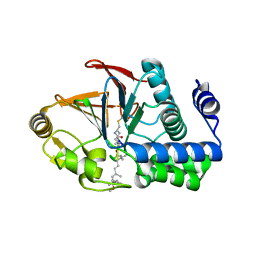 | | PP1 H66K in complex with Microcystin LR | | Descriptor: | MANGANESE (II) ION, Microcystin LR, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OFT
 
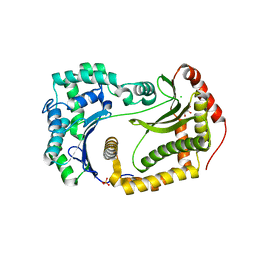 | |
6OBP
 
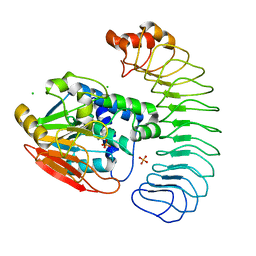 | | Reconstituted PP1 holoenzyme | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3BJF
 
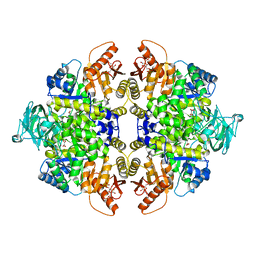 | | Pyruvate kinase M2 is a phosphotyrosine binding protein | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Wu, N. | | Deposit date: | 2007-12-03 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Pyruvate kinase M2 is a phosphotyrosine-binding protein.
Nature, 452, 2008
|
|
3AXL
 
 | | Murine Valpha 10 Vbeta 8.1 T-cell receptor | | Descriptor: | Valpha 10, Vbeta 8.1 | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2011-04-11 | | Release date: | 2011-08-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A semi-invariant V(alpha)10(+) T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties
Nat.Immunol., 12, 2011
|
|
3BJT
 
 | | Pyruvate kinase M2 is a phosphotyrosine binding protein | | Descriptor: | MAGNESIUM ION, OXALATE ION, Pyruvate kinase isozymes M1/M2 | | Authors: | Wu, N. | | Deposit date: | 2007-12-04 | | Release date: | 2008-03-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyruvate kinase M2 is a phosphotyrosine-binding protein.
Nature, 452, 2008
|
|
3BLS
 
 | | AMPC BETA-LACTAMASE FROM ESCHERICHIA COLI | | Descriptor: | AMPC BETA-LACTAMASE, M-AMINOPHENYLBORONIC ACID | | Authors: | Usher, K.C, Shoichet, B.K, Remington, S.J. | | Deposit date: | 1998-06-04 | | Release date: | 1998-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of AmpC beta-lactamase from Escherichia coli bound to a transition-state analogue: possible implications for the oxyanion hypothesis and for inhibitor design.
Biochemistry, 37, 1998
|
|
