1NYK
 
 | | Crystal Structure of the Rieske protein from Thermus thermophilus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, Rieske iron-sulfur protein | | Authors: | Hunsicker-Wang, L.M, Heine, A, Chen, Y, Luna, E.P, Todaro, T, Zhang, Y.M, Williams, P.A, McRee, D.E, Hirst, J, Stout, C.D, Fee, J.A. | | Deposit date: | 2003-02-12 | | Release date: | 2003-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High resolution structure of the soluble, respiratory-type Rieske protein from Thermus thermophilus: Analysis and Comparison
Biochemistry, 42, 2003
|
|
3PPP
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | Glycine betaine/carnitine/choline-binding protein, TRIMETHYL GLYCINE | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3LD0
 
 | | Crystal structure of B.licheniformis Anti-TRAP protein, an antagonist of TRAP-RNA interactions | | Descriptor: | Inhibitor of TRAP, regulated by T-BOX (Trp) sequence RtpA, MAGNESIUM ION, ... | | Authors: | Shevtsov, M.B, Chen, Y, Isupov, M.N, Gollnick, P, Antson, A.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bacillus licheniformis Anti-TRAP can assemble into two types of dodecameric particles with the same symmetry but inverted orientation of trimers.
J.Struct.Biol., 170, 2010
|
|
3ZZT
 
 | |
3PPO
 
 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (2S)-3-carboxy-2-hydroxy-N,N,N-trimethylpropan-1-aminium, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-24 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
3ZZU
 
 | |
3LRQ
 
 | | Crystal structure of the U-box domain of human ubiquitin-protein ligase (E3), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D. | | Descriptor: | E3 ubiquitin-protein ligase TRIM37, ZINC ION | | Authors: | Kuzin, A, Chen, Y, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-11 | | Release date: | 2010-03-23 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | E3 UBIQUITIN-PROTEIN LIGASE RING DOMAIN FROM HUMAN TRIPARTITE MOTIF-CONTAINING PROTEIN 37 (TRIM37), NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET HR4604D.
To be Published
|
|
3L9E
 
 | | Crystal structures of holo and Cu-deficient Cu/ZnSOD from the silkworm Bombyx mori and the implications in Amyotrophic lateral sclerosis | | Descriptor: | Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Zhang, N.-N, He, Y.-X, Li, W.-F, Zhao, F, Yan, L.-F, Zhang, G.-Z, Teng, Y.-B, Yu, J, Chen, Y, Zhou, C.-Z. | | Deposit date: | 2010-01-05 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of holo and Cu-deficient Cu/Zn-SOD from the silkworm Bombyx mori and the implications in amyotrophic lateral sclerosis
Proteins, 78, 2010
|
|
5H5U
 
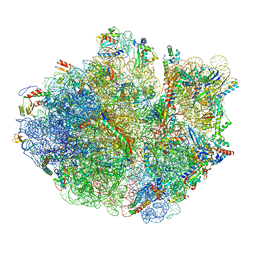 | | Mechanistic insights into the alternative translation termination by ArfA and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Ma, C, Kurita, D, Li, N, Chen, Y, Himeno, H, Gao, N. | | Deposit date: | 2016-11-09 | | Release date: | 2017-01-25 | | Last modified: | 2018-10-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic insights into the alternative translation termination by ArfA and RF2
Nature, 541, 2017
|
|
1M6X
 
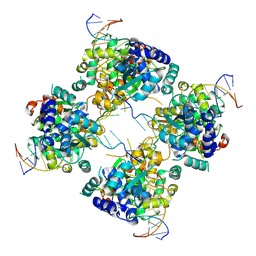 | | Flpe-Holliday Junction Complex | | Descriptor: | Flp recombinase, Symmetrized FRT site | | Authors: | Conway, A.B, Chen, Y, Rice, P.A. | | Deposit date: | 2002-07-17 | | Release date: | 2003-02-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Plasticity of the Flp-Holliday Junction Complex
J.Mol.Biol., 326, 2003
|
|
3LYX
 
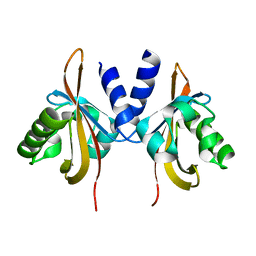 | | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B | | Descriptor: | Sensory box/GGDEF domain protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Janjua, H, Cunningham, K, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-02-28 | | Release date: | 2010-04-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the PAS domain of the protein CPS_1291 from Colwellia psychrerythraea. Northeast Structural Genomics Consortium target id CsR222B
To be Published
|
|
3PPR
 
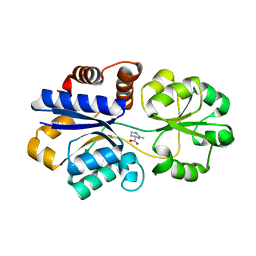 | | Structures of the substrate-binding protein provide insights into the multiple compatible solutes binding specificities of Bacillus subtilis ABC transporter OpuC | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, Glycine betaine/carnitine/choline-binding protein | | Authors: | Du, Y, Shi, W.W, He, Y.X, Yang, Y.H, Zhou, C.Z, Chen, Y. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC
Biochem.J., 436, 2011
|
|
4BVD
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 5-chloro-2-fluoro-N-[1-(4-piperidyl)pyrazol-4-yl]benzenesulfonamide, APOLIPOPROTEIN(A), CHLORIDE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BV7
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
1EV7
 
 | | CRYSTAL STRUCTURE OF DNA RESTRICTION ENDONUCLEASE NAEI | | Descriptor: | TYPE IIE RESTRICTION ENDONUCLEASE NAEI | | Authors: | Huai, Q, Colandene, J.D, Chen, Y, Luo, F, Zhao, Y. | | Deposit date: | 2000-04-19 | | Release date: | 2000-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of NaeI-an evolutionary bridge between DNA endonuclease and topoisomerase.
EMBO J., 19, 2000
|
|
4BVC
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-PIPERIDYL)-N-[2-(TRIFLUOROMETHOXY)PHENYL]SULFONYL-PROPANAMIDE, APOLIPOPROTEIN(A), CHLORIDE ION, ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
7CYE
 
 | | Cryo-EM structure of sodium-dependent bicarbonate transporter SbtA from Synechocystis sp. PCC 6803 | | Descriptor: | Slr1512 protein | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
7CYF
 
 | | Cryo-EM structure of bicarbonate transporter SbtA in complex with PII-like signaling protein SbtB from Synechocystis sp. PCC 6803 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION, ... | | Authors: | Liu, X.Y, Jiang, Y.L, Wang, L, Hou, W.T, Chen, Y, Zhou, C.Z. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-23 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures of cyanobacterial bicarbonate transporter SbtA and its complex with PII-like SbtB.
Cell Discov, 7, 2021
|
|
6L8L
 
 | | C-Src in complex with ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Characterization of ibrutinib as a non-covalent inhibitor of SRC-family kinases.
Bioorg.Med.Chem.Lett., 34, 2020
|
|
4F3L
 
 | | Crystal Structure of the Heterodimeric CLOCK:BMAL1 Transcriptional Activator Complex | | Descriptor: | BMAL1b, Circadian locomoter output cycles protein kaput | | Authors: | Huang, N, Chelliah, Y, Shan, Y, Taylor, C, Yoo, S, Partch, C, Green, C.B, Zhang, H, Takahashi, J. | | Deposit date: | 2012-05-09 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.268 Å) | | Cite: | Crystal structure of the heterodimeric CLOCK:BMAL1 transcriptional activator complex.
Science, 337, 2012
|
|
3MI6
 
 | | Crystal structure of the alpha-galactosidase from Lactobacillus brevis, Northeast Structural Genomics Consortium Target LbR11. | | Descriptor: | Alpha-galactosidase | | Authors: | Vorobiev, S, Chen, Y, Seetharaman, J, Belote, R, Sahdev, S, Xiao, R, Acton, T.B, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-04-09 | | Release date: | 2010-04-28 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of the alpha-galactosidase from Lactobacillus brevis.
To be Published
|
|
4BV5
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 4-(aminomethyl)-N-(benzenesulfonyl)cyclohexanecarboxamide, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
3MER
 
 | | Crystal Structure of the methyltransferase Slr1183 from Synechocystis sp. PCC 6803, Northeast Structural Genomics Consortium Target SgR145 | | Descriptor: | Slr1183 protein | | Authors: | Seetharaman, J, Chen, Y, Forouhar, F, Rossi, P, Sahdev, S, Xiao, R, Ciccosanti, C, Foote, E.L, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Northeast Structural Genomics Consortium Target SgR145
To be Published
|
|
4FES
 
 | | Structure of OSH4 in complex with cholesterol analogs | | Descriptor: | (3S,8S,9S,10R,13S,14S,17R)-3-hydroxy-10,13-dimethyl-17-[(2S,6S)-6-methyl-3-oxooctan-2-yl]-1,2,3,4,7,8,9,10,11,12,13,14,15,17-tetradecahydro-16H-cyclopenta[a]phenanthren-16-one, Protein KES1 | | Authors: | Koag, M.C, Monzingo, A.F, Cheun, Y, Lee, S. | | Deposit date: | 2012-05-30 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis and structure of 16,22-diketocholesterol bound to oxysterol-binding protein Osh4.
Steroids, 78, 2013
|
|
6L2W
 
 | | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1 | | Descriptor: | freshwater cyanophage protein | | Authors: | Wang, Y, Jin, H, Yang, F, Jiang, Y.L, Zhao, Y.Y, Chen, Z.P, Li, W.F, Chen, Y, Zhou, C.Z, Li, Q. | | Deposit date: | 2019-10-07 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of a novel fold protein Gp72 from the freshwater cyanophage Mic1.
Proteins, 88, 2020
|
|
