2GLP
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with compound 1 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2GLV
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) mutant(Y100A) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2GLL
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, BENZAMIDINE, CHLORIDE ION | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
2GLM
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(FabZ) from Helicobacter pylori complexed with Compound 2 | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 2-CHLORO-5-(5-{(E)-[(2Z)-3-(2-METHOXYETHYL)-4-OXO-2-(PHENYLIMINO)-1,3-THIAZOLIDIN-5-YLIDENE]METHYL}-2-FURYL)BENZOIC ACID, BENZAMIDINE, ... | | Authors: | Zhang, L, Liu, W, Shen, X, Jiang, H. | | Deposit date: | 2006-04-05 | | Release date: | 2007-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ).
J.Biol.Chem., 283, 2008
|
|
5EO3
 
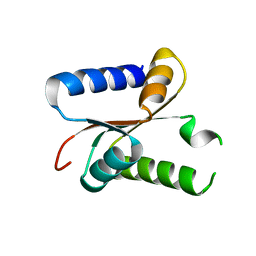 | |
8JNK
 
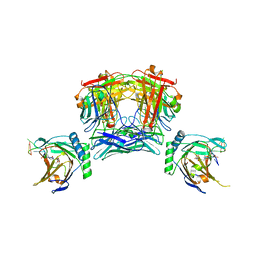 | |
8JNR
 
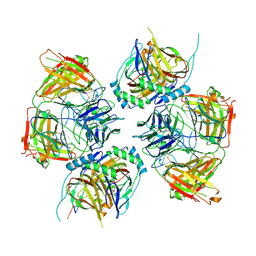 | |
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
8JKK
 
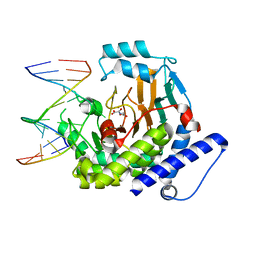 | |
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
6KVM
 
 | | Crystal structure of Chicken MHC Class II for 1.9 angstrom | | Descriptor: | GLYCEROL, MHC class II alpha chain, MHC class II beta chain 2, ... | | Authors: | Zhang, L, Xia, C. | | Deposit date: | 2019-09-04 | | Release date: | 2019-12-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | A Newly Recognized Pairing Mechanism of the alpha- and beta-Chains of the Chicken Peptide-MHC Class II Complex.
J Immunol., 204, 2020
|
|
6KXH
 
 | | Alp1U_Y247F mutant in complex with Fluostatin C | | Descriptor: | D-MALATE, Fluostatin C, Putative hydrolase, ... | | Authors: | Zhang, L, Yingli, Z, De, B.C, Zhang, C. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78039551 Å) | | Cite: | Mutation of an atypical oxirane oxyanion hole improves regioselectivity of the alpha / beta-fold epoxide hydrolase Alp1U.
J.Biol.Chem., 295, 2020
|
|
2C9R
 
 | | apo-H91F CopC | | Descriptor: | COPPER RESISTANCE PROTEIN C, SODIUM ION | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2C9P
 
 | | Cu(I)Cu(II)-CopC at pH 4.5 | | Descriptor: | COPPER (II) ION, COPPER RESISTANCE PROTEIN C, NITRATE ION | | Authors: | Zhang, L, Koay, M, Maher, M.J, Xiao, Z, Wedd, A.G. | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Intermolecular Transfer of Copper Ions from the Copc Protein of Pseudomonas Syringae. Crystal Structures of Fully Loaded Cu(I)Cu(II) Forms.
J.Am.Chem.Soc., 128, 2006
|
|
2FFK
 
 | |
2G0U
 
 | | Solution Structure of Monomeric BsaL, the Type III Secretion Needle Protein of Burkholderia pseudomallei | | Descriptor: | type III secretion system needle protein | | Authors: | Zhang, L, Wang, Y, Picking, W.L, Picking, W.D, De Guzman, R.N. | | Deposit date: | 2006-02-13 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Monomeric BsaL, the Type III Secretion Needle Protein of Burkholderia pseudomallei.
J.Mol.Biol., 359, 2006
|
|
2FIN
 
 | |
2H1Y
 
 | |
8YPY
 
 | |
8YPZ
 
 | |
8YQ0
 
 | |
3UOB
 
 | |
3UO7
 
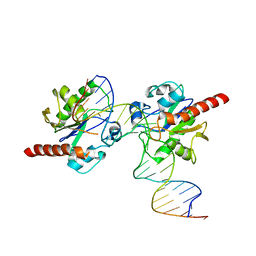 | | Crystal structure of Human Thymine DNA Glycosylase Bound to Substrate 5-carboxylcytosine | | Descriptor: | 5'-D(*CP*AP*GP*CP*TP*CP*TP*GP*TP*AP*CP*AP*TP*GP*AP*GP*CP*AP*GP*TP*GP*GP*A)-3', 5'-D(*CP*CP*AP*CP*TP*GP*CP*TP*CP*AP*(1CC)P*GP*TP*AP*CP*AP*GP*AP*GP*CP*TP*GP*T)-3', G/T mismatch-specific thymine DNA glycosylase | | Authors: | Zhang, L, He, C. | | Deposit date: | 2011-11-16 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Thymine DNA glycosylase specifically recognizes 5-carboxylcytosine-modified DNA.
Nat.Chem.Biol., 8, 2012
|
|
