7Z1N
 
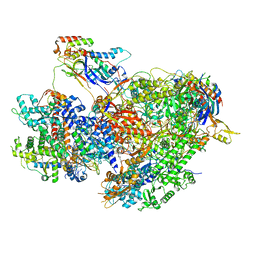 | | Structure of yeast RNA Polymerase III Delta C53-C37-C11 | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1O
 
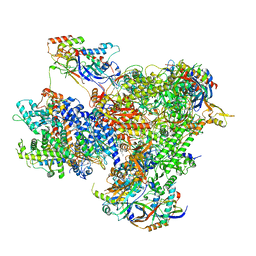 | | Structure of yeast RNA Polymerase III PTC + NTPs | | Descriptor: | CHAPSO, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
7Z1M
 
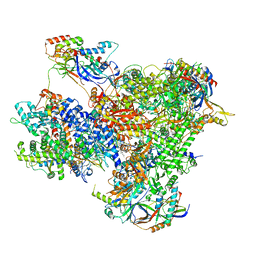 | | Structure of yeast RNA Polymerase III Elongation Complex (EC) | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, ... | | Authors: | Girbig, M, Mueller, C.W. | | Deposit date: | 2022-02-24 | | Release date: | 2022-08-31 | | Last modified: | 2022-09-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Architecture of the yeast Pol III pre-termination complex and pausing mechanism on poly(dT) termination signals.
Cell Rep, 40, 2022
|
|
4GWG
 
 | |
6NHQ
 
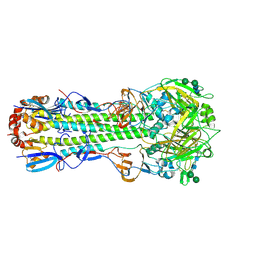 | |
6NHR
 
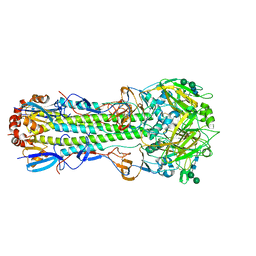 | |
6NHP
 
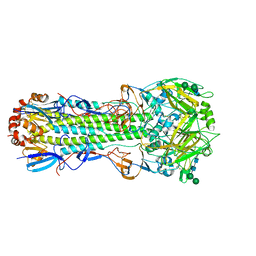 | |
7EMN
 
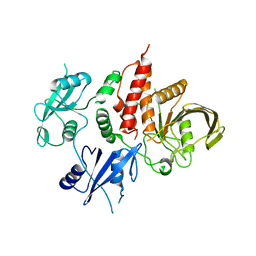 | | The atomic structure of SHP2 E76A mutant | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Luo, F, Xie, J.J, Zhu, J.D, Liu, C. | | Deposit date: | 2021-04-14 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A novel partially open state of SHP2 points to a "multiple gear" regulation mechanism.
J.Biol.Chem., 296, 2021
|
|
5UMN
 
 | |
5TGG
 
 | |
5TJD
 
 | | Computer-based rational design of improved functionality for antibody catalysts toward organophosphorus compounds | | Descriptor: | FAB A.17 L47K mutant HEAVY CHAIN, FAB A.17 L47K mutant Light Chain | | Authors: | Chatziefthimiou, S, Stepanova, A, Smirnov, I, Gabibov, A, Wilmanns, M. | | Deposit date: | 2016-10-04 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multiscale computation delivers organophosphorus reactivity and stereoselectivity to immunoglobulin scavengers.
Proc.Natl.Acad.Sci.USA, 2020
|
|
5VTR
 
 | |
5VU4
 
 | |
5VTW
 
 | |
2LZ3
 
 | |
2LZ4
 
 | |
2OQA
 
 | | X-ray Sequence and Crystal Structure of Luffaculin 1, a Novel Type 1 Ribosome-inactivating Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Luffaculin 1, ... | | Authors: | Hou, X, Huang, M. | | Deposit date: | 2007-01-31 | | Release date: | 2007-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray sequence and crystal structure of luffaculin 1, a novel type 1 ribosome-inactivating protein
Bmc Struct.Biol., 7, 2007
|
|
6BKQ
 
 | |
6BKO
 
 | |
6BKS
 
 | |
6BKT
 
 | |
6BKR
 
 | |
6BKM
 
 | |
6BKP
 
 | |
6BKN
 
 | |
