7ZUA
 
 | |
7ZV1
 
 | |
7ZTW
 
 | |
8A2F
 
 | | Crystal Structure of Ljunganvirus 1 2A protein | | Descriptor: | 2A protein | | Authors: | von Castelmur, E, Zhu, L, Wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family
To Be Published
|
|
8A2E
 
 | | Crystal Structure of Human Parechovirus 3 2A protein | | Descriptor: | 2A protein, GLYCEROL, SULFATE ION | | Authors: | von Castelmur, E, Zhu, L, wang, X, Fry, E, Ren, J, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family.
To Be Published
|
|
8A2G
 
 | | Crystal structure of Sebokelevirus 2A2 protein | | Descriptor: | 1,2-ETHANEDIOL, 2A2 protein, TETRAETHYLENE GLYCOL | | Authors: | Zhu, L, Von Castelmur, E, Whang, X, Ren, J, Fry, E, Perrakis, A, Stuart, D.I. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural plasticity of 2A proteins in the Parechovirus family
to be published
|
|
2X4Q
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a photocleavable peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Class I Major Histocompatibility Complexes Loaded by a Periodate Trigger.
J.Am.Chem.Soc., 131, 2009
|
|
2X4T
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a Peiodate- cleavable peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 65 KDA PHOSPHOPROTEIN, BETA-2-MICROGLOBULIN, ... | | Authors: | Rodenko, B, Toebes, M, Celie, P.H.N, Perrakis, A, Schumacher, T.N.M, Ovaa, H. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Class I Major Histocompatibility Complexes Loaded by a Periodate Trigger.
J.Am.Chem.Soc., 131, 2009
|
|
2X4P
 
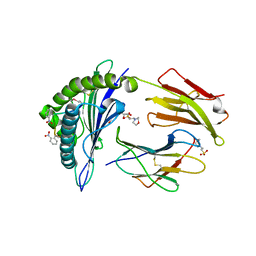 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a photocleavable peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Class I Major Histocompatibility Complexes Loaded by a Periodate Trigger.
J.Am.Chem.Soc., 131, 2009
|
|
8BBM
 
 | |
2XR9
 
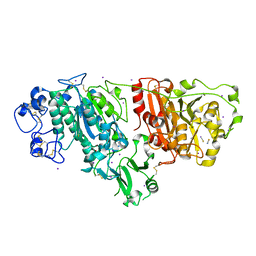 | | Crystal structure of Autotaxin (ENPP2) | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Kamtekar, S, Hausmann, J, Day, J.E, Christodoulou, E, Perrakis, A. | | Deposit date: | 2010-09-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of substrate discrimination and integrin binding by autotaxin.
Nat. Struct. Mol. Biol., 18, 2011
|
|
1EPU
 
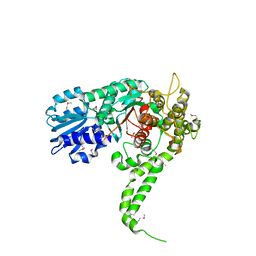 | | X-RAY crystal structure of neuronal SEC1 from squid | | Descriptor: | S-SEC1 | | Authors: | Bracher, A, Perrakis, A, Dresbach, T, Betz, H, Weissenhorn, W. | | Deposit date: | 2000-03-29 | | Release date: | 2000-08-09 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis.
Structure Fold.Des., 8, 2000
|
|
1O9K
 
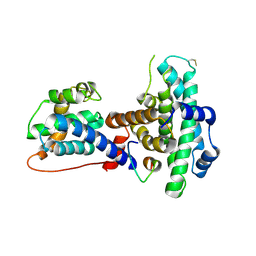 | | Crystal structure of the retinoblastoma tumour suppressor protein bound to E2F peptide | | Descriptor: | RETINOBLASTOMA-ASSOCIATED PROTEIN, TRANSCRIPTION FACTOR E2F1 | | Authors: | Xiao, B, Spencer, J, Clements, A, Ali-Khan, N, Mittnacht, S, Broceno, C, Burghammer, M, Perrakis, A, Marmorstein, R, Gamblin, S.J. | | Deposit date: | 2002-12-16 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Retinoblastoma Tumor Suppressor Protein Bound to E2F and the Molecular Basis of its Regulation
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1QBB
 
 | | BACTERIAL CHITOBIASE COMPLEXED WITH CHITOBIOSE (DINAG) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-07 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
1QBA
 
 | | BACTERIAL CHITOBIASE, GLYCOSYL HYDROLASE FAMILY 20 | | Descriptor: | CHITOBIASE, SULFATE ION | | Authors: | Tews, I, Perrakis, A, Oppenheim, A, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Bacterial chitobiase structure provides insight into catalytic mechanism and the basis of Tay-Sachs disease.
Nat.Struct.Biol., 3, 1996
|
|
2W8G
 
 | | Aplysia californica AChBP bound to in silico compound 35 | | Descriptor: | (3-ENDO,8-ANTI)-8-BENZYL-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
2W8F
 
 | | Aplysia californica AChBP bound to in silico compound 31 | | Descriptor: | (3-EXO)-3-(10,11-DIHYDRO-5H-DIBENZO[A,D][7]ANNULEN-5-YLOXY)-8,8-DIMETHYL-8-AZONIABICYCLO[3.2.1]OCTANE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Ulens, C, Akdemir, A, Jongejan, A, van Elk, R, Edink, E, Bertrand, S, Perrakis, A, Leurs, R, Smit, A.B, Sixma, T.K, Bertrand, D, de Esch, I.J. | | Deposit date: | 2009-01-16 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Use of Acetylcholine Binding Protein in the Search for Novel Alpha7 Nicotinic Receptor Ligands. In Silico Docking, Pharmacological Screening, and X- Ray Analysis.
J.Med.Chem., 52, 2009
|
|
2WVR
 
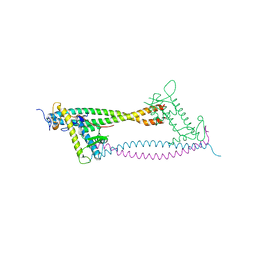 | | Human Cdt1:Geminin complex | | Descriptor: | DNA REPLICATION FACTOR CDT1, GEMININ | | Authors: | De Marco, V, Perrakis, A. | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Quaternary Structure of the Human Cdt1-Geminin Complex Regulates DNA Replication Licensing.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2X4O
 
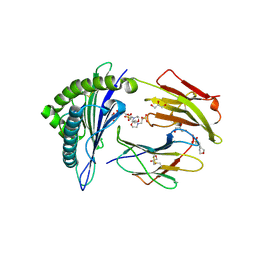 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 envelope peptide env120-128 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, ENVELOPE GLYCOPROTEIN GP160, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2X70
 
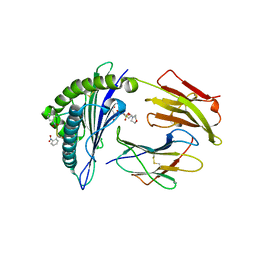 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a photocleavable peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-22 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2XSE
 
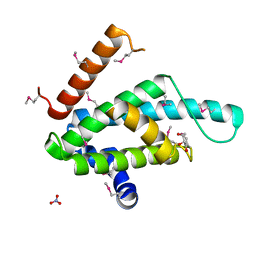 | | The structural basis for recognition of J-base containing DNA by a novel DNA-binding domain in JBP1 | | Descriptor: | GLYCEROL, NITRATE ION, THYMINE DIOXYGENASE JBP1 | | Authors: | Heidebrecht, T, Christodoulou, E, Chalmers, M.J, Jan, S, ter Riete, B, Grover, R.K, Joosten, R.P, Littler, D, vanLuenen, H, Griffin, P.R, Wentworth, P, Borst, P, Perrakis, A. | | Deposit date: | 2010-09-28 | | Release date: | 2011-03-30 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structural Basis for Recognition of Base J Containing DNA by a Novel DNA Binding Domain in Jbp1.
Nucleic Acids Res., 39, 2011
|
|
2XRG
 
 | | Crystal structure of Autotaxin (ENPP2) in complex with the HA155 boronic acid inhibitor | | Descriptor: | CALCIUM ION, ECTONUCLEOTIDE PYROPHOSPHATASE/PHOSPHODIESTERASE FAMILY MEMBER 2, IODIDE ION, ... | | Authors: | Hausmann, J, Albers, H.M.H.G, Perrakis, A. | | Deposit date: | 2010-09-14 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Basis of Substrate Discrimination and Integrin Binding by Autotaxin.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2X4N
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to residual fragments of a photocleavable peptide that is cleaved upon UV-light treatment | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2X4U
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to HIV-1 Peptide RT468-476 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
2X4R
 
 | | Crystal structure of MHC CLass I HLA-A2.1 bound to Cytomegalovirus (CMV) pp65 epitope | | Descriptor: | 65 KDA PHOSPHOPROTEIN, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | Authors: | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | Deposit date: | 2010-02-02 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
