8GY6
 
 | |
6AAX
 
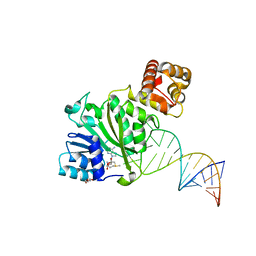 | | Crystal structure of TFB1M and h45 with SAM in homo sapiens | | Descriptor: | DI(HYDROXYETHYL)ETHER, Dimethyladenosine transferase 1, mitochondrial, ... | | Authors: | Liu, X, Shen, S, Wu, P, Li, F, Gong, Q, Wu, J, Zhang, H, Shi, Y. | | Deposit date: | 2018-07-19 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.994 Å) | | Cite: | Structural insights into dimethylation of 12S rRNA by TFB1M: indispensable role in translation of mitochondrial genes and mitochondrial function.
Nucleic Acids Res., 47, 2019
|
|
8HOK
 
 | | crystal structure of UGT71AP2 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, UGT71AP2 | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'- O -glycosyltransferase from Scutellaria baicalensis .
Acta Pharm Sin B, 14, 2024
|
|
8HOJ
 
 | | Crystal structure of UGT71AP2 in complex with UDP | | Descriptor: | UGT71AP2, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, Z.L, He, C, Li, F, Qiao, X, Ye, M. | | Deposit date: | 2022-12-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional characterization, structural basis, and protein engineering of a rare flavonoid 2'- O -glycosyltransferase from Scutellaria baicalensis .
Acta Pharm Sin B, 14, 2024
|
|
4ZRJ
 
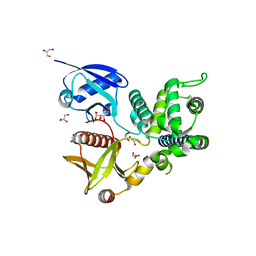 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4JGV
 
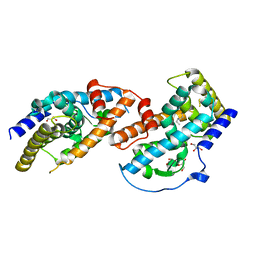 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
7R1T
 
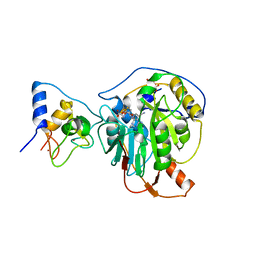 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
7R1U
 
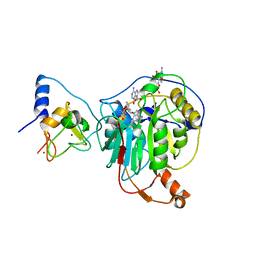 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
4GXL
 
 | | The crystal structure of Galectin-8 C-CRD in complex with NDP52 | | Descriptor: | GLYCEROL, Galectin-8, Peptide from Calcium-binding and coiled-coil domain-containing protein 2 | | Authors: | Li, S, Wandel, M.P, Li, F, Liu, Z, He, C, Wu, J, Shi, Y, Randow, F. | | Deposit date: | 2012-09-04 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.023 Å) | | Cite: | Sterical hindrance promotes selectivity of the autophagy cargo receptor NDP52 for the danger receptor galectin-8 in antibacterial autophagy
Sci.Signal., 6, 2013
|
|
6ACV
 
 | | the solution NMR structure of MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 11 | | Authors: | Li, S.L, Feng, Y.Y, Zhou, Y, Ding, Y.M, Liu, K, Nie, Y, Li, F, Yang, Y.Y. | | Deposit date: | 2018-07-27 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | the solution NMR structure of MBD domains
To Be Published
|
|
8HKD
 
 | |
8THF
 
 | | SARS-CoV-2 BA.1 S-6P-no-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1,Spike glycoprotein | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-07-15 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Universal subunit vaccine protects against multiple SARS-CoV-2 variants and SARS-CoV.
Npj Vaccines, 9, 2024
|
|
7UXE
 
 | | Pseudomonas phage E217 small terminase (TerS) | | Descriptor: | Small terminase | | Authors: | Lokareddy, R.K, Hou, C.-F.D, Doll, S.G, Li, F, Gillilan, R, Forti, F, Briani, F, Cingolani, G. | | Deposit date: | 2022-05-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Terminase Subunits from the Pseudomonas-Phage E217.
J.Mol.Biol., 434, 2022
|
|
5JI8
 
 | | Crystal structure of the BRD9 bromodomain and hit 1 | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, Bromodomain-containing protein 9 | | Authors: | Wang, N, Li, F, Bao, H, Li, J, Wu, J, Ruan, K. | | Deposit date: | 2016-04-22 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | NMR Fragment Screening Hit Induces Plasticity of BRD7/9 Bromodomains
Chembiochem, 17, 2016
|
|
8UPX
 
 | | Omicron-S-MERS-RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,SARS-CoV-2 omicron spike with MERS RBD | | Authors: | Bu, F, Li, F, Liu, B. | | Deposit date: | 2023-10-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Pan-beta-coronavirus subunit vaccine prevents SARS-CoV-2 Omicron, SARS-CoV, and MERS-CoV challenge.
J.Virol., 98, 2024
|
|
8UG9
 
 | | XBB.1.5 spike/Nb5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-10-05 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Dual-role epitope on SARS-CoV-2 spike enhances and neutralizes viral entry across different variants.
Plos Pathog., 20, 2024
|
|
8G7B
 
 | |
8G70
 
 | | SARS-CoV-2 spike/nanobody mixture complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 spike/nanobody mixture complex
To Be Published
|
|
8G76
 
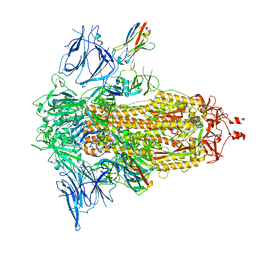 | | SARS-CoV-2 spike/Nb5 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Dual-role epitope on SARS-CoV-2 spike enhances and neutralizes viral entry across different variants.
Plos Pathog., 20, 2024
|
|
8G77
 
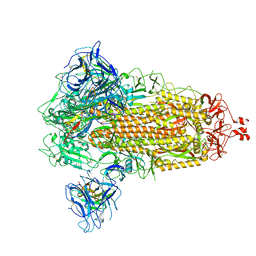 | | SARS-CoV-2 spike/Nb6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-6, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Dual-role epitope on SARS-CoV-2 spike enhances and neutralizes viral entry across different variants.
Plos Pathog., 20, 2024
|
|
8G7A
 
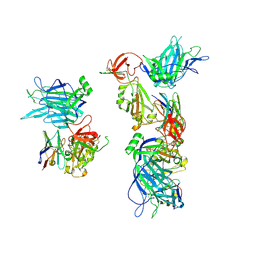 | |
8G7C
 
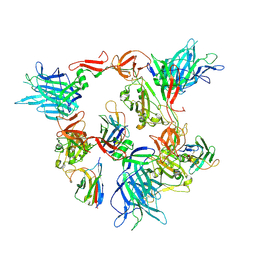 | |
8G78
 
 | | Local refinement of SARS-CoV-2 spike/nanobody mixture complex around NTD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-5, Nanosota-6, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Local refinement of SARS-CoV-2 spike/nanobody mixture complex around NTD
To Be Published
|
|
8G72
 
 | | SARS-CoV-2 spike/Nb2 complex with 1 RBD up (local refinement at 5.6 A) | | Descriptor: | Nanosota-2, Spike glycoprotein | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G74
 
 | | SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
