6MZF
 
 | |
8HGH
 
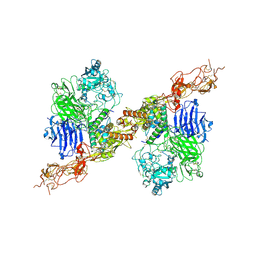 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
8HGG
 
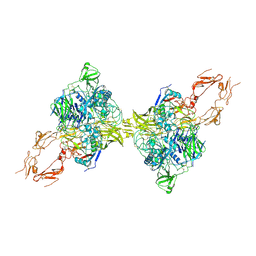 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
1NTY
 
 | |
4ZRI
 
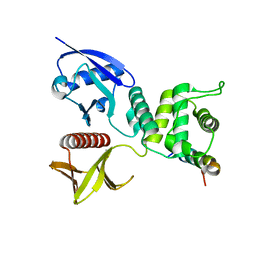 | | Crystal structure of Merlin-FERM and Lats2 | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS2 | | Authors: | Li, F, Zhou, H, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4ZRJ
 
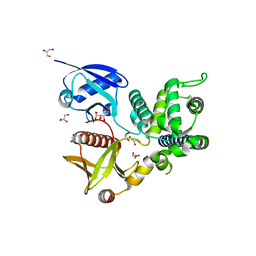 | | Structure of Merlin-FERM and CTD | | Descriptor: | GLYCEROL, Merlin | | Authors: | Lin, Z, Li, F, Long, J, Shen, Y. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
4ZRK
 
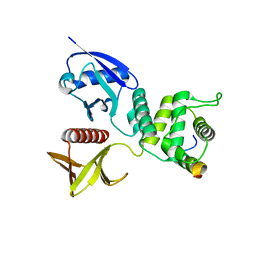 | | Merlin-FERM and Lats1 complex | | Descriptor: | Merlin, Serine/threonine-protein kinase LATS1 | | Authors: | Lin, Z, Li, Y, Wei, Z, Zhang, M. | | Deposit date: | 2015-05-12 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.316 Å) | | Cite: | Angiomotin binding-induced activation of Merlin/NF2 in the Hippo pathway
Cell Res., 25, 2015
|
|
5K8L
 
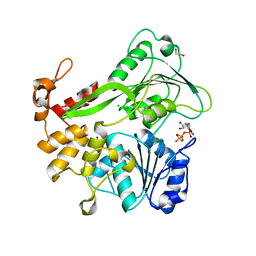 | | Crystal structure of ZIKV NS3 helicase in complex with GTP-gammar S | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5K8U
 
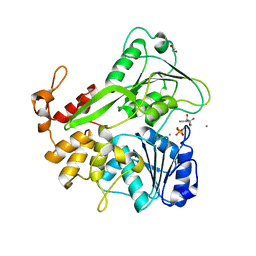 | | Crystal structure of ZIKV NS3 helicase in complex with ADP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-31 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5K8I
 
 | | Crystal structure of ZIKV NS3 helicase in complex with ATP and Mn2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.694 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5K8T
 
 | | Crystal structure of ZIKV NS3 helicase in complex with GTP-gammar S and an magnesium ion | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-31 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.848 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
5JWH
 
 | | Apo structure | | Descriptor: | 1,2-ETHANEDIOL, NS3 helicase | | Authors: | Cao, X, Li, Y, Jin, T. | | Deposit date: | 2016-05-12 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design.
Nucleic Acids Res., 44, 2016
|
|
7LUX
 
 | | AALALL segment from the Nucleoprotein of SARS-CoV-2, residues 217-222, crystal form 2 | | Descriptor: | Nucleoprotein AALALL, TETRAETHYLENE GLYCOL | | Authors: | Lu, J, Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.303 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
7LUZ
 
 | |
7LV2
 
 | |
7LTU
 
 | | AALALL SEGMENT FROM THE NUCLEOPROTEIN OF SARS-COV-2, RESIDUES 217-222, CRYSTAL FORM 1 | | Descriptor: | AALALL SEGMENT FROM THE NUCLEOPROTEIN OF SARS-COV-2,RESIDUES 217-222, trifluoroacetic acid | | Authors: | Zee, C.-T, Sawaya, M.R, Rodriguez, J.A, Eisenberg, D.S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-03-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.122 Å) | | Cite: | Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2.
Biorxiv, 2021
|
|
3DBO
 
 | | Crystal structure of a member of the VapBC family of toxin-antitoxin systems, VapBC-5, from Mycobacterium tuberculosis | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Miallau, L, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI), TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structure and Proposed Activity of a Member of the VapBC Family of Toxin-Antitoxin Systems: VapBC-5 FROM MYCOBACTERIUM TUBERCULOSIS.
J.Biol.Chem., 284, 2009
|
|
6D8M
 
 | |
6D8O
 
 | |
6D8N
 
 | |
6D8L
 
 | |
2HOI
 
 | |
2HOF
 
 | |
1KBU
 
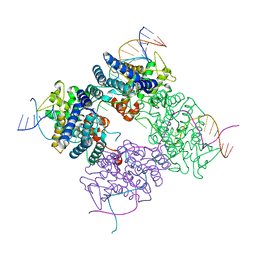 | | CRE RECOMBINASE BOUND TO A LOXP HOLLIDAY JUNCTION | | Descriptor: | CRE RECOMBINASE, LOXP | | Authors: | Martin, S.S, Pulido, E, Chu, V.C, Lechner, T, Baldwin, E.P. | | Deposit date: | 2001-11-06 | | Release date: | 2002-06-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Order of Strand Exchanges in Cre-LoxP Recombination and its Basis Suggested by the Crystal Structure of a
Cre-LoxP Holliday Junction Complex
J.Mol.Biol., 319, 2002
|
|
5WPW
 
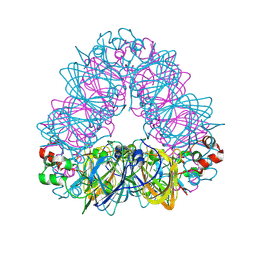 | | Crystal structure of coconut allergen cocosin | | Descriptor: | 11S globulin isoform 1 | | Authors: | Jin, T, Zhang, Y. | | Deposit date: | 2016-11-21 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Crystal Structure of Cocosin, A Potential Food Allergen from Coconut (Cocos nucifera)
J. Agric. Food Chem., 65, 2017
|
|
