5UZK
 
 | | Crystal Structure of PKA bound to an pyrrolo pyridine inhibitor | | Descriptor: | 2-{3-[3-(piperidin-4-yl)propoxy]phenyl}-N-[4-(1H-pyrrolo[2,3-b]pyridin-3-yl)-1,3-thiazol-2-yl]acetamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Jacobs, M.D, Brown, K. | | Deposit date: | 2017-02-26 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ROCK inhibitors 3: Design, synthesis and structure-activity relationships of 7-azaindole-based Rho kinase (ROCK) inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
8WP5
 
 | | Crystal structure of LbUGT1 in complex with UDP | | Descriptor: | CHLORIDE ION, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Hu, D, Wang, G.Q. | | Deposit date: | 2023-10-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.57005954 Å) | | Cite: | Functional and structural dissection of glycosyltransferases underlying the glycodiversity of wolfberry-derived bioactive ingredients lycibarbarspermidines.
Nat Commun, 15, 2024
|
|
8W53
 
 | | Crystal structure of LbUGT in complex with UDP | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Hu, D, Wang, G.Q. | | Deposit date: | 2023-08-25 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.42816162 Å) | | Cite: | Functional and structural dissection of glycosyltransferases underlying the glycodiversity of wolfberry-derived bioactive ingredients lycibarbarspermidines.
Nat Commun, 15, 2024
|
|
7ERO
 
 | | Crystal structure of D-allulose 3-epimerase with D-allulose from Agrobacterium sp. SUL3 | | Descriptor: | D-psicose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
7ERN
 
 | | Crystal structure of D-allulose 3-epimerase with D-fructose from Agrobacterium sp. SUL3 | | Descriptor: | D-fructose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
7ERM
 
 | | Crystal structure of D-allulose 3-epimerase from Agrobacterium sp. SUL3 | | Descriptor: | D-tagatose 3-epimerase, MAGNESIUM ION, SULFATE ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-05-06 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Growth-Coupled Evolutionary Pressure Improving Epimerases for D-Allulose Biosynthesis Using a Biosensor-Assisted In Vivo Selection Platform
Adv Sci, 2024
|
|
8J5Z
 
 | | The cryo-EM structure of the TwOSC1 tetramer | | Descriptor: | Terpene cyclase/mutase family member, octyl beta-D-glucopyranoside | | Authors: | Ma, X, Yuru, T, Yunfeng, L, Jiang, T. | | Deposit date: | 2023-04-24 | | Release date: | 2023-11-01 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (4.75 Å) | | Cite: | Structural and Catalytic Insight into the Unique Pentacyclic Triterpene Synthase TwOSC.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8SWV
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody IF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IF1 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8SWX
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody Base4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Base4 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8SWW
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody IF3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NHP polyclonal antibody IF3 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ward, A.B. | | Deposit date: | 2023-05-19 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8T2E
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FP3 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8T2F
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody N289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N289 Heavy Chain, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-06-05 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Priming antibody responses to the fusion peptide in rhesus macaques.
Npj Vaccines, 9, 2024
|
|
8SW7
 
 | | BG505 Boost2 SOSIP.664 in complex with NHP polyclonal antibody FP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 Boost 2 gp120, BG505 Boost 2 gp41, ... | | Authors: | Pratap, P.P, Antansijevic, A, Ozorowski, G, Ward, A.B. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Focusing antibody responses to the fusion peptide in rhesus macaques.
Biorxiv, 2023
|
|
3G9N
 
 | |
3G9L
 
 | |
3G90
 
 | |
3IO7
 
 | | 2-Aminopyrazolo[1,5-a]pyrimidines as potent and selective inhibitors of JAK2 | | Descriptor: | (3S)-1-[6-(2-aminopyrazolo[1,5-a]pyrimidin-3-yl)pyrimidin-4-yl]-N,N-diethylpiperidine-3-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Ledeboer, M.W, Pierce, A.C. | | Deposit date: | 2009-08-13 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Aminopyrazolo[1,5-a]pyrimidines as potent and selective inhibitors of JAK2.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IOK
 
 | | 2-Aminopyrazolo[1,5-a]pyrimidines as potent and selective inhibitors of JAK2 | | Descriptor: | 3-(6-{[(1S)-1-(4-fluorophenyl)ethyl]amino}pyrimidin-4-yl)pyrazolo[1,5-a]pyrimidin-2-amine, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Ledeboer, M.W, Pierce, A.C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-11-10 | | Last modified: | 2013-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2-Aminopyrazolo[1,5-a]pyrimidines as potent and selective inhibitors of JAK2.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
5BML
 
 | | ROCK 1 bound to a pyridine thiazole inhibitor | | Descriptor: | N-[4-(2-fluoropyridin-4-yl)thiophen-2-yl]-2-{3-[(methylsulfonyl)amino]phenyl}acetamide, Rho-associated protein kinase 1 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Design, Synthesis, and Structure-Activity Relationships of Pyridine-Based Rho Kinase (ROCK) Inhibitors.
J.Med.Chem., 58, 2015
|
|
2GV9
 
 | |
4NCM
 
 | | Influenza polymerase basic protein 2 (PB2) bound to a small-molecule inhibitor | | Descriptor: | N~2~-[2-(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-fluoropyrimidin-4-yl]-N,N-dimethyl-L-alaninamide, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of a Novel, First-in-Class, Orally Bioavailable Azaindole Inhibitor (VX-787) of Influenza PB2.
J.Med.Chem., 57, 2014
|
|
4NCE
 
 | | Influenza polymerase basic protein 2 (PB2) bound to 7-methyl-GTP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, 9,10-dioxo-9,10-dihydroanthracene-2,6-disulfonic acid, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2013-10-24 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of a Novel, First-in-Class, Orally Bioavailable Azaindole Inhibitor (VX-787) of Influenza PB2.
J.Med.Chem., 57, 2014
|
|
5F79
 
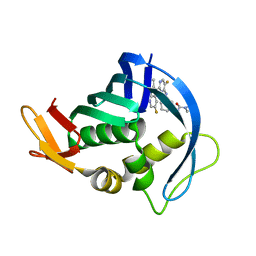 | | Influenza PB2 bound to an azaindole inhibitor | | Descriptor: | N-[(1R,3S)-3-({5-fluoro-2-[5-fluoro-2-(hydroxymethyl)-1H-pyrrolo[2,3-b]pyridin-3-yl]pyrimidin-4-yl}amino)cyclohexyl]pyrrolidine-1-carboxamide, Polymerase basic protein 2 | | Authors: | Jacobs, M.D. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel 2-Substituted 7-Azaindole and 7-Azaindazole Analogues as Potential Antiviral Agents for the Treatment of Influenza.
ACS Med Chem Lett, 8, 2017
|
|
5FA5
 
 | |
4P1U
 
 | |
