4LYE
 
 | |
3GZX
 
 | | Crystal Structure of the Biphenyl Dioxygenase in complex with Biphenyl from Comamonas testosteroni Sp. Strain B-356 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BIPHENYL, Biphenyl dioxygenase subunit alpha, ... | | Authors: | Kumar, P, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2009-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Characterization of Pandoraea pnomenusa B-356 Biphenyl Dioxygenase Reveals Features of Potent Polychlorinated Biphenyl-Degrading Enzymes
Plos One, 8, 2013
|
|
3GZY
 
 | | Crystal Structure of the Biphenyl Dioxygenase from Comamonas testosteroni Sp. Strain B-356 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Biphenyl dioxygenase subunit alpha, Biphenyl dioxygenase subunit beta, ... | | Authors: | Kumar, P, Colbert, C.L, Bolin, J.T. | | Deposit date: | 2009-04-08 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Characterization of Pandoraea pnomenusa B-356 Biphenyl Dioxygenase Reveals Features of Potent Polychlorinated Biphenyl-Degrading Enzymes
Plos One, 8, 2013
|
|
1LKD
 
 | | CRYSTAL STRUCTURE OF 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE (DHBD) COMPLEXED WITH 2',6'-DICL DIHYDROXYBIPHENYL (DHB) | | Descriptor: | 2',6'-DICHLORO-BIPHENYL-2,6-DIOL, BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE, FE (II) ION, ... | | Authors: | Dai, S, Bolin, J.T. | | Deposit date: | 2002-04-24 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and analysis of a bottleneck in PCB biodegradation
Nat.Struct.Biol., 9, 2002
|
|
1LGT
 
 | | CRYSTAL STRUCTURE OF 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE (DHBD) COMPLEXED WITH 2'-Cl DIHYDROXYBIPHENYL (DHB) | | Descriptor: | 2'-CHLORO-BIPHENYL-2,3-DIOL, BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE, FE (II) ION | | Authors: | Dai, S, Bolin, J.T. | | Deposit date: | 2002-04-16 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and analysis of a bottleneck in PCB biodegradation
Nat.Struct.Biol., 12, 2002
|
|
5CXG
 
 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional repressor KstR, TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1001 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5CYV
 
 | | Crystal structure of CouR from Rhodococcus jostii RHA1 bound to p-coumaroyl-CoA | | Descriptor: | ACETATE ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Stogios, P.J, Xu, X, Dong, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The activity of CouR, a MarR family transcriptional regulator, is modulated through a novel molecular mechanism.
Nucleic Acids Res., 44, 2016
|
|
4LXI
 
 | | Crystal Structure of the S105A mutant of a carbon-carbon bond hydrolase, DxnB2 from Sphingomonas wittichii RW1, in complex with 5,8-diF HOPDA | | Descriptor: | (3E,5R)-5-fluoro-6-(2-fluorophenyl)-2,6-dioxohex-3-enoic acid, MCP Hydrolase, SODIUM ION | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A substrate-assisted mechanism of nucleophile activation in a ser-his-asp containing C-C bond hydrolase.
Biochemistry, 52, 2013
|
|
8VBW
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Ertapenem) inhibited form | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBU
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Oxacillin) inhibited form | | Descriptor: | (2R,4S)-5,5-dimethyl-2-[(1R)-1-{[(5-methyl-3-phenyl-1,2-oxazol-4-yl)carbonyl]amino}-2-oxoethyl]-1,3-thiazolidine-4-carb oxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBV
 
 | | Structure of the monofunctional Staphylococcus aureus PBP1 in its beta-lactam (Cephalexin) inhibited form | | Descriptor: | (2S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5-methyl-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 1 | | Authors: | Bon, C.G, Lee, J, Caveney, N.A, Strynadka, N.C.J. | | Deposit date: | 2023-12-12 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and kinetic analysis of the monofunctional Staphylococcus aureus PBP1.
J.Struct.Biol., 216, 2024
|
|
8VBT
 
 | |
2PU5
 
 | | Crystal Structure of a C-C bond hydrolase, BphD, from Burkholderia xenovorans LB400 | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2PUH
 
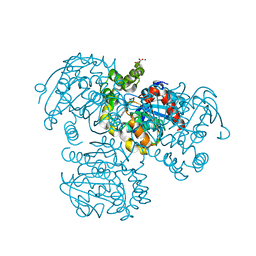 | | Crystal Structure of the S112A mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, in complex with its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, ... | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2PU7
 
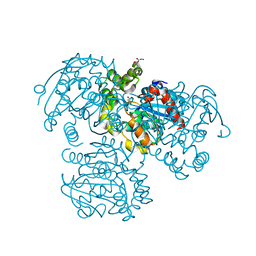 | | Crystal Structure of S112A/H265A double mutant of a C-C hydrolase, BphD, from Burkholderia xenovorans LB400 | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, SODIUM ION | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2PUJ
 
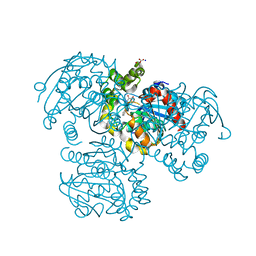 | | Crystal Structure of the S112A/H265A double mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, in complex with its substrate HOPDA | | Descriptor: | (2E,4E)-2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOIC ACID, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, ... | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2RI6
 
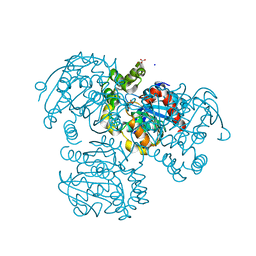 | | Crystal Structure of S112A mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400 | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, SODIUM ION | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-10-10 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The tautomeric half-reaction of BphD, a C-C bond hydrolase. Kinetic and structural evidence supporting a key role for histidine 265 of the catalytic triad.
J.Biol.Chem., 282, 2007
|
|
6C3D
 
 | | O2-, PLP-dependent L-arginine hydroxylase RohP quinonoid II complex | | Descriptor: | (2E,3E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pent-3-enoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
6C3B
 
 | | O2-, PLP-Dependent L-Arginine Hydroxylase RohP Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
6C39
 
 | |
6C3C
 
 | | PLP-dependent L-arginine hydroxylase RohP quinonoid I complex | | Descriptor: | (2E)-5-carbamimidamido-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}pentanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
6C3K
 
 | |
6C3A
 
 | | O2-, PLP-dependent L-arginine hydroxylase RohP 4-hydroxy-2-ketoarginine complex | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
5TW4
 
 | |
5TX9
 
 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with ceftobiprole | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyr rolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Penicillin-binding protein 4, SODIUM ION, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-16 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
