4AXX
 
 | |
4C4T
 
 | |
4C4R
 
 | |
5OK2
 
 | |
5OK1
 
 | |
5OK0
 
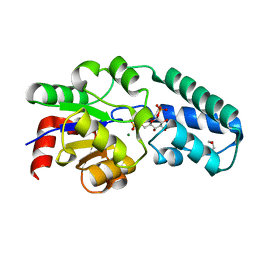 | | Structure of the D10N mutant of beta-phosphoglucomutase from Lactococcus lactis trapped with native reaction intermediate beta-glucose 1,6-bisphosphate to 2.2A resolution. | | Descriptor: | 1,3-PROPANDIOL, 1,6-di-O-phosphono-beta-D-glucopyranose, Beta-phosphoglucomutase, ... | | Authors: | Robertson, A.J, Bisson, C. | | Deposit date: | 2017-07-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | van der Waals Contact between Nucleophile and Transferring Phosphorus Is Insufficient To Achieve Enzyme Transition-State Architecture
Acs Catalysis, 2018
|
|
5O6R
 
 | |
5OJZ
 
 | |
5O6P
 
 | |
7NRJ
 
 | |
7O5T
 
 | |
7O5N
 
 | | Crystal Structure of a Class D carbapenemase complexed with Avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1-BUTANOL, Beta-lactamase, ... | | Authors: | Zhou, Q, Zhang, Z, He, Y, Jin, Y. | | Deposit date: | 2021-04-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
7O9N
 
 | |
7PGO
 
 | | Crystal Structure of a Class D Carbapenemase_R250A | | Descriptor: | 1-BUTANOL, BROMIDE ION, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
7PFN
 
 | |
7PEH
 
 | | Crystal Structure of a Class D Carbapenemase | | Descriptor: | 1-BUTANOL, Beta-lactamase | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
7PEP
 
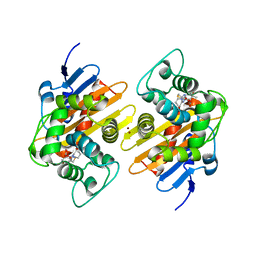 | | Crystal Structure of a Class D Carbapenemase Complexed with Hydrolyzed Imipenem | | Descriptor: | (2R,4S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-[(2-{[(Z)-iminomethyl]amino}ethyl)sulfanyl]-3,4-dihydro-2H-pyrrole-5-ca rboxylic acid, 1-BUTANOL, BROMIDE ION, ... | | Authors: | Zhou, Q, He, Y, Jin, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An Ion-Pair Induced Intermediate Complex Captured in Class D Carbapenemase Reveals Chloride Ion as a Janus Effector Modulating Activity
Acs Cent.Sci., 2023
|
|
7PEI
 
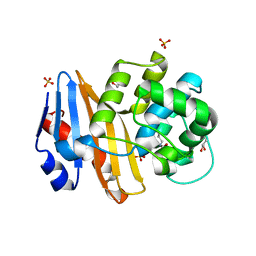 | |
16PK
 
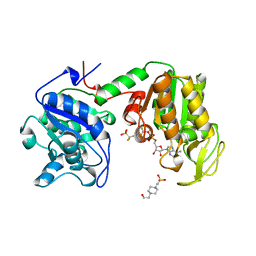 | | PHOSPHOGLYCERATE KINASE FROM TRYPANOSOMA BRUCEI BISUBSTRATE ANALOG | | Descriptor: | 1,1,5,5-TETRAFLUOROPHOSPHOPENTYLPHOSPHONIC ACID ADENYLATE ESTER, 3-PHOSPHOGLYCERATE KINASE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID | | Authors: | Bernstein, B.E, Bressi, J, Blackburn, M, Gelb, M, Hol, W.G.J. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bisubstrate analog induces unexpected conformational changes in phosphoglycerate kinase from Trypanosoma brucei.
J.Mol.Biol., 279, 1998
|
|
2XE6
 
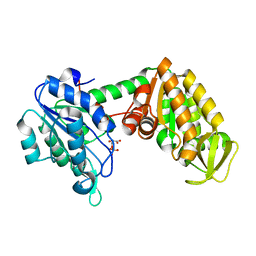 | | The complete reaction cycle of human phosphoglycerate kinase: The open binary complex with 3PG | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
2XE8
 
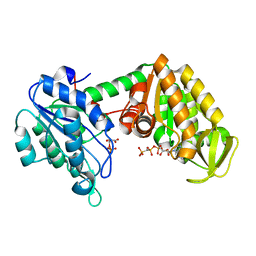 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and AMP-PNP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PHOSPHOGLYCERATE KINASE 1, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
2XE7
 
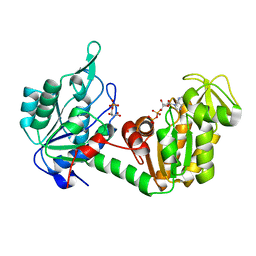 | | The complete reaction cycle of human phosphoglycerate kinase: The open ternary complex with 3PG and ADP | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
2F1D
 
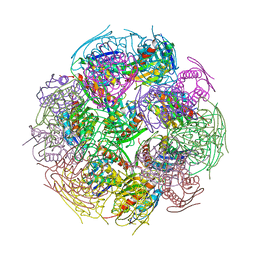 | | X-Ray Structure of imidazoleglycerol-phosphate dehydratase | | Descriptor: | Imidazoleglycerol-phosphate dehydratase 1, MANGANESE (II) ION, SULFATE ION | | Authors: | Rice, D.W, Glynn, S.E, Baker, P.J, Sedelnikova, S.E, Davies, C.L, Eadsforth, T.C. | | Deposit date: | 2005-11-14 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and mechanism of imidazoleglycerol-phosphate dehydratase.
Structure, 13, 2005
|
|
2H2J
 
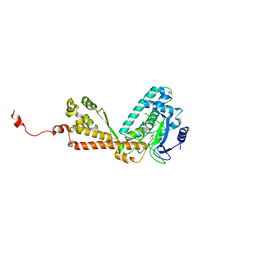 | | Structure of Rubisco LSMT bound to Sinefungin and Monomethyllysine | | Descriptor: | N-METHYL-LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, SINEFUNGIN | | Authors: | Couture, J.F, Hauk, G, Trievel, R.C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Catalytic Roles for Carbon-Oxygen Hydrogen Bonding in SET Domain Lysine Methyltransferases.
J.Biol.Chem., 281, 2006
|
|
2H2E
 
 | | Structure of Rubisco LSMT bound to AzaAdoMet and Lysine | | Descriptor: | LYSINE, Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE | | Authors: | Couture, J.F, Hauk, G, Trievel, R.C. | | Deposit date: | 2006-05-18 | | Release date: | 2006-05-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Catalytic Roles for Carbon-Oxygen Hydrogen Bonding in SET Domain Lysine Methyltransferases.
J.Biol.Chem., 281, 2006
|
|
