7B0P
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus in space group P21212 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, GLYCEROL, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7B0Q
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus with H85A mutation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CITRIC ACID, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
7B0R
 
 | | In meso structure of the membrane integral lipoprotein intramolecular transacylase Lit from Bacillus cereus with H85R mutation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Huang, C.-Y, Olatunji, S, Olieric, V, Caffrey, M. | | Deposit date: | 2020-11-20 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of the membrane intramolecular transacylase reaction responsible for lyso-form lipoprotein synthesis.
Nat Commun, 12, 2021
|
|
6FMV
 
 | | IMISX-EP of Hg-BacA soaking SIRAS | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMX
 
 | | IMISX-EP of W-PgpB | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphatidylglycerophosphatase B, TUNGSTATE(VI)ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMR
 
 | | IMISX-EP of Se-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMW
 
 | | IMISX-EP of Hg-BacA cocrystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMT
 
 | | IMISX-EP of Hg-BacA Soaking SAD | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6FMY
 
 | | IMISX-EP of S-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Di-or tripeptide:H+ symporter, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
4BPM
 
 | | Crystal structure of a human integral membrane enzyme | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, PROSTAGLANDIN E SYNTHASE, ... | | Authors: | Li, D, Wang, M, Olieric, V, Caffrey, M. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystallizing Membrane Proteins in the Lipidic Mesophase. Experience with Human Prostaglandin E2 Synthase 1 and an Evolving Strategy.
Cryst.Growth Des., 14, 2014
|
|
6FMS
 
 | | IMISX-EP of Se-LspA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Globomycin, Lipoprotein signal peptidase | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
4NC3
 
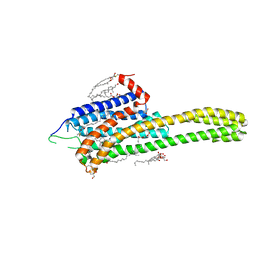 | | Crystal structure of the 5-HT2B receptor solved using serial femtosecond crystallography in lipidic cubic phase. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHOLESTEROL, ... | | Authors: | Liu, W, Wacker, D, Gati, C, Han, G.W, James, D, Wang, D, Nelson, G, Weierstall, U, Katritch, V, Barty, A, Zatsepin, N.A, Li, D, Messerschmidt, M, Boutet, S, Williams, G.J, Koglin, J.E, Seibert, M.M, Wang, C, Shah, S.T.A, Basu, S, Fromme, R, Kupitz, C, Rendek, K.N, Grotjohann, I, Fromme, P, Kirian, R.A, Beyerlein, K.R, White, T.A, Chapman, H.N, Caffrey, M, Spence, J.C.H, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serial femtosecond crystallography of G protein-coupled receptors.
Science, 342, 2013
|
|
1WJB
 
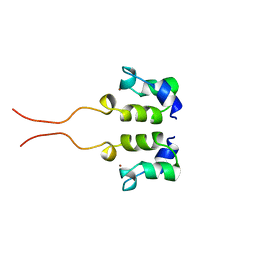 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, 40 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJC
 
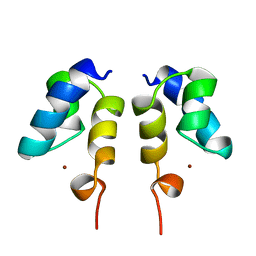 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJA
 
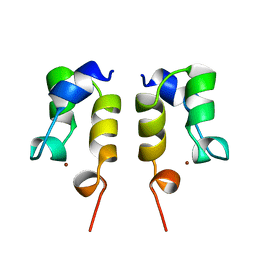 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (D FORM), NMR, REGULARIZED MEAN STRUCTURE | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
1WJD
 
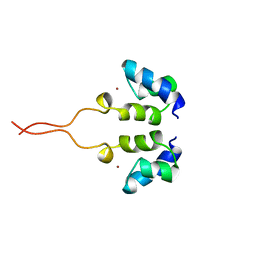 | | SOLUTION STRUCTURE OF THE N-TERMINAL ZN BINDING DOMAIN OF HIV-1 INTEGRASE (E FORM), NMR, 38 STRUCTURES | | Descriptor: | HIV-1 INTEGRASE, ZINC ION | | Authors: | Clore, G.M, Cai, M, Caffrey, M, Gronenborn, A.M. | | Deposit date: | 1997-05-13 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc binding domain of HIV-1 integrase.
Nat.Struct.Biol., 4, 1997
|
|
2VDF
 
 | | Structure of the OpcA adhesion from Neisseria meningitidis determined by crystallization from the cubic mesophase | | Descriptor: | N-OCTANE, OUTER MEMBRANE PROTEIN, SULFATE ION | | Authors: | Cherezov, V, Liu, W, Derrick, J.P, Luan, B, Aksimentiev, A, Katritch, V, Caffrey, M. | | Deposit date: | 2007-10-05 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In meso crystal structure and docking simulations suggest an alternative proteoglycan binding site in the OpcA outer membrane adhesin.
Proteins, 71, 2008
|
|
5D59
 
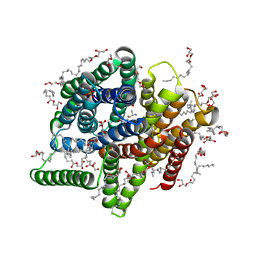 | | In meso X-ray crystallography structure of the PepTSt-Ala-Phe complex at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ALANINE, ... | | Authors: | Huang, C.-Y, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5C
 
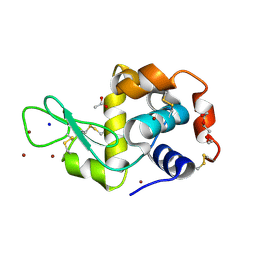 | | In meso in situ serial X-ray crystallography structure of lysozyme at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D52
 
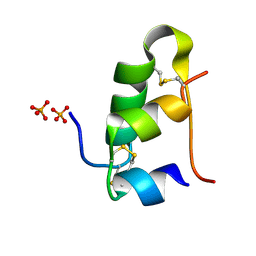 | | In meso in situ serial X-ray crystallography structure of insulin at room temperature | | Descriptor: | Insulin A chain, Insulin B chain, PHOSPHATE ION | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D54
 
 | | In meso X-ray crystallography structure of insulin at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Insulin A chain, Insulin B chain, ... | | Authors: | Huang, C.-Y, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D57
 
 | | In meso X-ray crystallography structure of diacylglycerol kinase, DgkA, at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Diacylglycerol kinase | | Authors: | Huang, C.-Y, Howe, N, Olieric, V, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5E
 
 | | In meso in situ serial X-ray crystallography structure of insulin by sulfur-SAD at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Insulin A chain, Insulin B chain, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5A
 
 | | In meso in situ serial X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Liu, X, Kobilka, B, Kay Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4826 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5F
 
 | | In meso in situ serial X-ray crystallography structure of lysozyme by bromine-SAD at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ACETIC ACID, BROMIDE ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2016-03-02 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
