8VKN
 
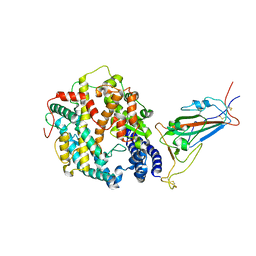 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (focused refinement of RBD and mouse ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
7N83
 
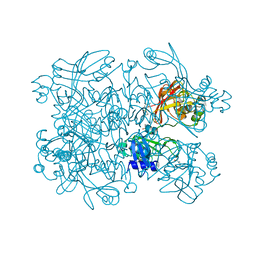 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z2443429438 | | Descriptor: | (3S)-1-(phenylsulfonyl)pyrrolidin-3-amine, CITRIC ACID, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-12 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
8V7U
 
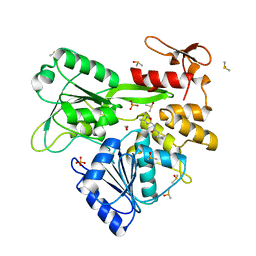 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclopentyl-N-(3-methyl-1,2,4-oxadiazol-5-yl)acetamide, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z729726784
To Be Published
|
|
8UIP
 
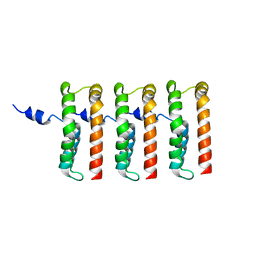 | |
8WO7
 
 | | Apo state of Arabidopsis AZG1 T440Y | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Adenine/guanine permease AZG1 | | Authors: | Xu, L, Guo, J. | | Deposit date: | 2023-10-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and mechanisms of the Arabidopsis cytokinin transporter AZG1.
Nat.Plants, 10, 2024
|
|
8VKL
 
 | | Cryo-EM structure of SARS-CoV-2 XBB.1.5 spike protein in complex with mouse ACE2 (conformation 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J, Poloni, C, Bezeruk, A, Tidey, K, Ahmed, S, Tuttle, K, Vahdatihassani, F, Cholak, S, Cook, L, Steiner, T.S, Subramaniam, S. | | Deposit date: | 2024-01-09 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | SARS-CoV-2 XBB.1.5 Spike Protein: Altered Receptor Binding, Antibody Evasion, and Retention of T Cell Recognition
To Be Published
|
|
9AVA
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2R)-2-[(5R,6S,8R,9aS)-8-amino-1-oxo-5-(2-phenylethyl)hexahydro-1H-pyrrolo[1,2-a][1,4]diazepin-2(3H)-yl]-N-[(3,4-dichlorophenyl)methyl]-4-methylpentanamide, POTASSIUM ION, Three-prime repair exonuclease 1, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Xu, J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-01 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9F4X
 
 | | UP1 in complex with Z1220452176 | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed, ~{N}-[2-(5-fluoranyl-1~{H}-indol-3-yl)ethyl]ethanamide | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
5KWL
 
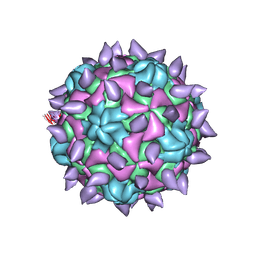 | | expanded poliovirus in complex with VHH 10E | | Descriptor: | VHH 10E, VP1, VP2, ... | | Authors: | Strauss, M, Schotte, L, Filman, D.J, Hogle, J.M. | | Deposit date: | 2016-07-18 | | Release date: | 2016-11-02 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-electron Microscopy Structures of Expanded Poliovirus with VHHs Sample the Conformational Repertoire of the Expanded State.
J. Virol., 91, 2017
|
|
7N7U
 
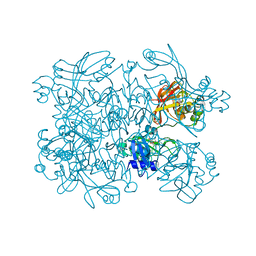 | | Crystal Structure of SARS-CoV-2 NendoU in complex with LIZA-7 | | Descriptor: | 1-[(2~{R},4~{S},5~{R})-5-[[(azanylidene-$l^{4}-azanylidene)amino]methyl]-4-oxidanyl-oxolan-2-yl]-5-methyl-pyrimidine-2,4-dione, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
8Z90
 
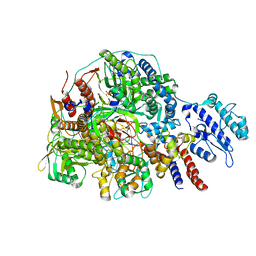 | | Cryo-EM structure of Thogoto virus polymerase in transcription initiation conformation 2 | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
9F4Q
 
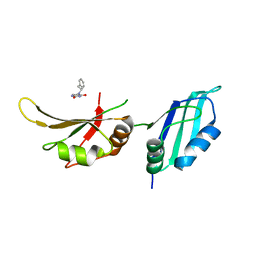 | | UP1 in complex with Z641239276 | | Descriptor: | 5-(2-phenylethylamino)pyrimidine-2,4-diol, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
8Z98
 
 | | Cryo-EM structure of Thogoto virus polymerase in a transcription reception conformation | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8ZJV
 
 | | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Yang, Y, Fu, L, Chen, R, Cheng, H, Sun, X. | | Deposit date: | 2024-05-15 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the ERK2 complexed with 5-Iodotubercidin
To Be Published
|
|
8UWP
 
 | | Crystal structure of SETDB1 Tudor domain in complex with MR46747 | | Descriptor: | (3S)-N-(4-chloro-3-{[2-(diethylamino)ethyl]carbamoyl}phenyl)-3-(diethylamino)pyrrolidine-1-carboxamide, 1,2-ETHANEDIOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Shrestha, S, Beldar, S, Dong, A, Ackloo, S, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-07 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with MR46747
To be published
|
|
5ALD
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 5-cyclohexyl-1,3-dihydroindol-2-one, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
8TKA
 
 | |
9F54
 
 | | UP1 in complex with EN300-197154 | | Descriptor: | 1-methyl-3-[(3R)-piperidin-3-yl]-1H-pyrazole-4-carboxamide, Heterogeneous nuclear ribonucleoprotein A1, N-terminally processed | | Authors: | Dunnett, L, Prischi, F. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Enhanced identification of small molecules binding to hnRNP A1 via in silico hotspot and cryptic pockets mapping coupled with X-Ray fragment screening
To Be Published
|
|
7N7R
 
 | | Crystal Structure of SARS-CoV-2 NendoU in complex with Z2472938267 | | Descriptor: | 1-[2-(2-oxidanylidenepyrrolidin-1-yl)ethyl]-3-phenyl-urea, Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Douangamath, A, Nakamura, A.M, Dias, A, Krojer, T, Noske, G.D, Gawiljuk, V.O, Fernandes, R.S, Fairhead, M, Powell, A, Dunnet, L, Aimon, A, Fearon, D, Brandao-Neto, J, Skyner, R, von Delft, F, Oliva, G. | | Deposit date: | 2021-06-11 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
5AIA
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 5-cyclohexyl-2-hydroxybenzoate, BIFUNCTIONAL EPOXIDE HYDROLASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-02-12 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
8UD0
 
 | |
8TL8
 
 | | Structure of Orthoreovirus RNA Chaperone SigmaNS R6A mutant in complex with bile acid | | Descriptor: | GLYCOCHOLIC ACID, Protein sigma-NS | | Authors: | Prasad, B.V.V, Zhao, B, Hu, L, Neetu, N. | | Deposit date: | 2023-07-26 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of orthoreovirus RNA chaperone sigma NS, a component of viral replication factories.
Nat Commun, 15, 2024
|
|
5AJS
 
 | |
8Y66
 
 | | Cryo-EM structure of human urate transporter GLUT9 bound to inhibitor apigenin | | Descriptor: | 5,7-dihydroxy-2-(4-hydroxyphenyl)-4H-chromen-4-one, Solute carrier family 2, facilitated glucose transporter member 9 | | Authors: | Pan, X.J, Shen, Z.L, Xu, L, Huang, G.X.Y. | | Deposit date: | 2024-02-01 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for urate recognition and apigenin inhibition of human GLUT9.
Nat Commun, 15, 2024
|
|
5KYD
 
 | |
