8G1Z
 
 | |
8G20
 
 | |
7DZ9
 
 | | MbnABC complex | | Descriptor: | FE (III) ION, MbnA, MbnB, ... | | Authors: | Chao, D, Dan, Z, Yijun, G, Wei, C. | | Deposit date: | 2021-01-25 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
7NUS
 
 | |
5MAG
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5NB7
 
 | | Complement factor D | | Descriptor: | 1-[2-[(1~{R},3~{S},5~{R})-3-[(6-bromanylpyridin-2-yl)carbamoyl]-2-azabicyclo[3.1.0]hexan-2-yl]-2-oxidanylidene-ethyl]indazole-3-carboxamide, Complement factor D, DIMETHYL SULFOXIDE | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5NAR
 
 | |
5NAW
 
 | | Complement factor D in complex with the inhibitor (1R,3S,5R)-2-Aza-bicyclo[3.1.0]hexane-2,3-dicarboxylic acid 2-[(1-carbamoyl-1H-indol-3-yl)-amide] 3-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (1~{R},3~{S},5~{R})-~{N}2-(1-aminocarbonylindol-3-yl)-~{N}3-[3-(trifluoromethyloxy)phenyl]-2-azabicyclo[3.1.0]hexane-2,3-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-02-28 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
5M5A
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, K-252A, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-10-21 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5MAH
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5MAF
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Maternal embryonic leucine zipper kinase, ... | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5MAI
 
 | | Crystal structure of MELK in complex with an inhibitor | | Descriptor: | 3-[(3~{Z})-3-[[[4-[(dimethylamino)methyl]phenyl]amino]-phenyl-methylidene]-2-oxidanylidene-1~{H}-indol-6-yl]-~{N}-ethyl-prop-2-ynamide, DIMETHYL SULFOXIDE, Maternal embryonic leucine zipper kinase | | Authors: | Canevari, G, Re Depaolini, S, Casale, E, Felder, E, Kuster, B, Heinzlmeir, S. | | Deposit date: | 2016-11-03 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The target landscape of clinical kinase drugs.
Science, 358, 2017
|
|
5NBA
 
 | | Complement factor D in complex with the inhibitor (2S,4R)-4-Fluoro-pyrrolidine-1,2-dicarboxylic acid 1-[(1-carbamoyl-1H-indol-3-yl)-amide] 2-[(3-trifluoromethoxy-phenyl)-amide] | | Descriptor: | (2~{S},4~{R})-~{N}1-(1-aminocarbonylindol-3-yl)-4-fluoranyl-~{N}2-[3-(trifluoromethyloxy)phenyl]pyrrolidine-1,2-dicarboxamide, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N. | | Deposit date: | 2017-03-01 | | Release date: | 2017-06-28 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Highly Potent and Selective Small-Molecule Reversible Factor D Inhibitors Demonstrating Alternative Complement Pathway Inhibition in Vivo.
J. Med. Chem., 60, 2017
|
|
8IYG
 
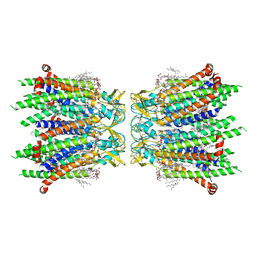 | | Human neuronal gap junction channel connexin 36 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, Gap junction delta-2 protein | | Authors: | Mao, W.X, Chen, S.S. | | Deposit date: | 2023-04-04 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Assembly mechanisms of the neuronal gap junction channel connexin 36 elucidated by Cryo-EM.
Arch.Biochem.Biophys., 754, 2024
|
|
8ANF
 
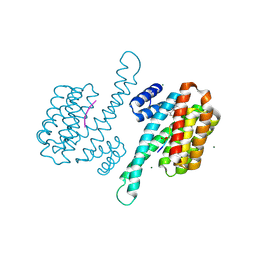 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074359) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[3-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]propyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-05 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AXU
 
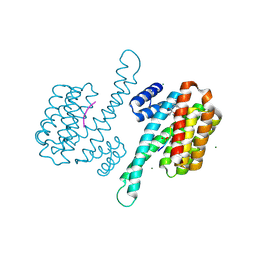 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075297) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-(4-chloranylphenoxy)cyclopentyl]carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AWG
 
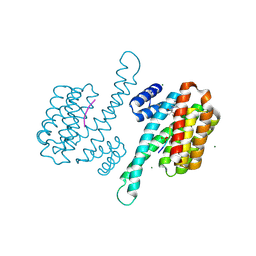 | | small molecule stabilizer for ERalpha and 14-3-3 (1074202) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-29 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AZE
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075306) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-[1-[(4-chlorophenyl)amino]cyclopentyl]carbonylpiperidin-4-yl]methyl]ethanamide, ERalpha peptide, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AOY
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1075478) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-N-[3-[1-[4-[(4-chlorophenyl)amino]oxan-4-yl]carbonylpiperidin-4-yl]propyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-09 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8AXE
 
 | | Small molecule stabilizer for ERalpha and 14-3-3 (1074210) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[2-[1-[2-(4-chloranylphenoxy)-2-methyl-propanoyl]piperidin-4-yl]ethyl]ethanamide, Estrogen receptor, ... | | Authors: | Konstantinidou, M, Visser, E.J, Vandenboorn, E.M.F, Sheng, C, Jaishankar, P, Overmans, M.J.A.M, Dutta, S, Neitz, J, Renslo, A, Ottmann, C, Brunsveld, L, Arkin, M. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
7FC0
 
 | | Reconstitution of MbnABC complex from Rugamonas rubra ATCC-43154 (GroupIII) | | Descriptor: | FE (III) ION, Methanobactin biosynthesis cassette protein MbnB, Methanobactin biosynthesis cassette protein MbnC, ... | | Authors: | Chao, D, Zhaolin, L, Shoujie, L, Li, Z, Dan, Z, Ying, J, Wei, C. | | Deposit date: | 2021-07-13 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.643 Å) | | Cite: | Crystal structure and catalytic mechanism of the MbnBC holoenzyme required for methanobactin biosynthesis.
Cell Res., 32, 2022
|
|
6IR1
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM4 at 1.9 Angstron resolution | | Descriptor: | MCherry fluorescent protein, mCherry's nanobody LaM4 | | Authors: | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6E66
 
 | |
8HFR
 
 | | NPC-trapped pre-60S particle | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-11-12 | | Release date: | 2023-04-12 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
8HBN
 
 | | Structure of the Mex67-Mtr2-1 heterodimer | | Descriptor: | mRNA export factor MEX67, mRNA transport regulator MTR2 | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-10-29 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
