6IZQ
 
 | | PRMT4 bound with a bicyclic compound | | Descriptor: | (2R)-1-(methylamino)-3-(1,3,4,5-tetrahydro-2-benzazepin-2-yl)propan-2-ol, Histone-arginine methyltransferase CARM1 | | Authors: | Xiong, B, Cao, D.Y, Guo, Z.H, Li, Y.L, Li, J, Huang, X, Shen, J.K. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Design and Synthesis of Potent, Selective Inhibitors of Protein Arginine Methyltransferase 4 against Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
8JVA
 
 | | Cryo-EM structure of the N-terminal domain of Omicron BA.1 in complex with nanobody N235 and S2L20 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S2L20 heavy chain, S2L20 light chain, ... | | Authors: | Liu, B, Liu, H.H, Han, P, Qi, J.X. | | Deposit date: | 2023-06-28 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Enhanced potency of an IgM-like nanobody targeting conserved epitope in SARS-CoV-2 spike N-terminal domain.
Signal Transduct Target Ther, 9, 2024
|
|
9I02
 
 | | Structure of recombinant human butyrylcholinesterase in complex with (S)-N-((1-benzylpyrrolidin-3-yl)methyl)-N-methylnaphthalene-2-sulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Kosak, U, Gobec, S, Nachon, F. | | Deposit date: | 2025-01-14 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Lead Optimization of a Butyrylcholinesterase Inhibitor for the Treatment of Alzheimer's Disease.
J.Med.Chem., 68, 2025
|
|
9I03
 
 | | Structure of recombinant human butyrylcholinesterase in complex with (R)-N-((1-benzylpyrrolidin-3-yl)methyl)-N-methylnaphthalene-2-sulfonamide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Kosak, U, Gobec, S, Nachon, F. | | Deposit date: | 2025-01-14 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Lead Optimization of a Butyrylcholinesterase Inhibitor for the Treatment of Alzheimer's Disease.
J.Med.Chem., 68, 2025
|
|
5ZE3
 
 | |
6JGY
 
 | | Crystal structure of LASV-GP2 in a post fusion conformation | | Descriptor: | Pre-glycoprotein polyprotein GP complex | | Authors: | Zhu, Y, Zhang, X, Chen, B, Ye, S, Zhang, R. | | Deposit date: | 2019-02-15 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.389 Å) | | Cite: | Crystal Structure of Refolding Fusion Core of Lassa Virus GP2 and Design of Lassa Virus Fusion Inhibitors.
Front Microbiol, 10, 2019
|
|
9IYQ
 
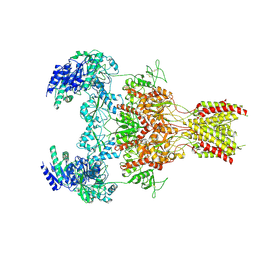 | | Structure of the human GluN1-N2B NMDA receptors in the Mg2+ free state | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Huang, X, Sun, X, Zhu, S. | | Deposit date: | 2024-07-31 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into the diverse actions of magnesium on NMDA receptors.
Neuron, 113, 2025
|
|
9IYP
 
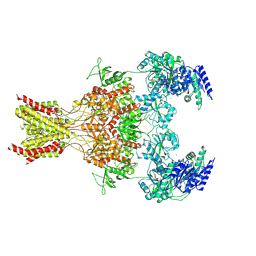 | | Structure of the human GluN1-N2B NMDA receptors in the Mg2+ bound state | | Descriptor: | (2R)-4-(3-phosphonopropyl)piperazine-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Huang, X, Sun, X, Zhu, S. | | Deposit date: | 2024-07-31 | | Release date: | 2025-03-05 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the diverse actions of magnesium on NMDA receptors.
Neuron, 113, 2025
|
|
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
9J3C
 
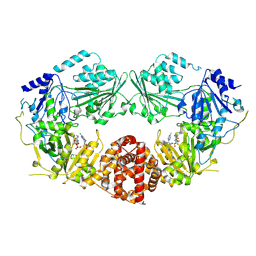 | | Cryo-EM structure of NAT10 with Co-enzyme A | | Descriptor: | COENZYME A, RNA cytidine acetyltransferase | | Authors: | Jiang, Y, Xia, J. | | Deposit date: | 2024-08-08 | | Release date: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Targeting NAT10 attenuates homologous recombination via destabilizing DNA:RNA hybrids and overcomes PARP inhibitor resistance in cancers.
Drug Resist Updat., 81, 2025
|
|
7DHX
 
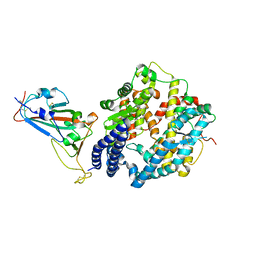 | | Crystal structure of SARS-CoV-2 RBD binding to pangolin ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ZINC ION, ... | | Authors: | Wang, Q.H, Qi, J.X, Wu, L.L. | | Deposit date: | 2020-11-17 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of pangolin ACE2 engaged by COVID-19 virus
Chin.Sci.Bull., 66, 2021
|
|
8IGA
 
 | |
4AV4
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to a propynyl pyridine | | Descriptor: | 3-(pyridin-3-yl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-23 | | Release date: | 2012-06-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Tyrosine Gate as a Potential Entropic Lever in the Receptor-Binding Site of the Bacterial Adhesin Fimh.
Biochemistry, 51, 2012
|
|
4AUJ
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to para hydroxypropargyl phenyl | | Descriptor: | 4-(3-hydroxyprop-1-yn-1-yl)phenyl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-17 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.527 Å) | | Cite: | Validation of Reactivity Descriptors to Assess the Aromatic Stacking within the Tyrosine Gate of Fimh.
Acs Med.Chem.Lett., 4, 2013
|
|
8JRN
 
 | | Structure of E6AP-E6 complex in Att1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRQ
 
 | | Structure of E6AP-E6 complex in Det1 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRP
 
 | | Structure of E6AP-E6 complex in Att3 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRR
 
 | | Structure of E6AP-E6 complex in Det2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
8JRO
 
 | | Structure of E6AP-E6 complex in Att2 state | | Descriptor: | Protein E6, Ubiquitin-protein ligase E3A, ZINC ION | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2023-06-17 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into the functional mechanism of the ubiquitin ligase E6AP.
Nat Commun, 15, 2024
|
|
4ATT
 
 | | FimH lectin domain co-crystal with a alpha-D-mannoside O-linked to a propynyl para methoxy phenyl | | Descriptor: | 3-(4-methoxyphenyl)prop-2-yn-1-yl alpha-D-mannopyranoside, FIMH | | Authors: | Bouckaert, J, Touaibia, M, Roos, G, Shiao, T.C, Wang, Q, Papadopoulos, A, Roy, R. | | Deposit date: | 2012-05-09 | | Release date: | 2013-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.251 Å) | | Cite: | Validation of Reactivity Descriptors to Assess the Aromatic Stacking within the Tyrosine Gate of Fimh.
Acs Med.Chem.Lett., 4, 2013
|
|
8Y86
 
 | | Human AE3 with NaHCO3- | | Descriptor: | Anion exchange protein 3, BICARBONATE ION | | Authors: | Jian, L, Zhang, Q, Yao, D, Cao, Y. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | The structural insight into the functional modulation of human anion exchanger 3.
Nat Commun, 15, 2024
|
|
8XM2
 
 | |
7EIN
 
 | | SARS-CoV-2 main proteinase complex with microbial metabolite leupeptin | | Descriptor: | 3C-like proteinase, leupeptin | | Authors: | Fu, L.F, Feng, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Microbial Metabolite Leupeptin in the Treatment of COVID-19 by Traditional Chinese Medicine Herbs.
Mbio, 12, 2021
|
|
5Z9O
 
 | |
8IG4
 
 | | Crystal structure of SARS-Cov-2 main protease in complex with GC376 | | Descriptor: | Non-structural protein 11, N~2~-[(benzyloxy)carbonyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Coronaviral Main Proteases Inhibition by the 3CLpro Inhibitor GC376.
J.Mol.Biol., 436, 2024
|
|
