6WFG
 
 | | Crystal structure of human Naa50 in complex with an inhibitor (compound 3) identified using DNA encoded library technology | | Descriptor: | (2S)-N-[(2S)-3-[1-(3-tert-butyl-1-methyl-1H-pyrazole-5-carbonyl)piperidin-4-yl]-1-(methylamino)-1-oxopropan-2-yl]-6-oxopiperidine-2-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFO
 
 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4b) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5R)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WF3
 
 | | Crystal structure of human Naa50 in complex with a cofactor derived inhibitor (compound 1) | | Descriptor: | ACE-MET-LEU-GLY-PRO-NH2, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFN
 
 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFK
 
 | | Crystal structure of human Naa50 in complex with CoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
7YBO
 
 | | Crystal structure of FGFR4 kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC3
 
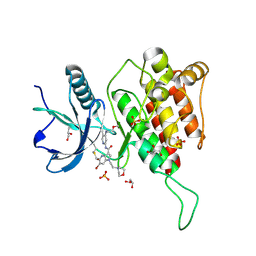 | | Crystal structure of FGFR4 kinase domain with 10t | | Descriptor: | 6-bromanyl-~{N}-[5-cyano-4-(2-methoxyethylamino)pyridin-2-yl]-5-methanoyl-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide, Fibroblast growth factor receptor 4, GLYCEROL, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.987 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YC1
 
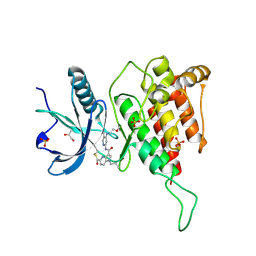 | | Crystal structure of FGFR4 kinase domain with 10d | | Descriptor: | Fibroblast growth factor receptor 4, GLYCEROL, SULFATE ION, ... | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.535 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7YBP
 
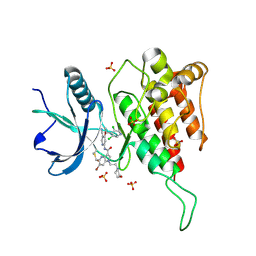 | | Crystal structure of FGFR4(V550L) kinase domain with 10z | | Descriptor: | Fibroblast growth factor receptor 4, SULFATE ION, ~{N}-[4-[(1~{R})-1-[3,5-bis(chloranyl)pyridin-4-yl]ethoxy]-5-cyano-pyridin-2-yl]-6-bromanyl-5-(hydroxymethyl)-1-(2-morpholin-4-ylethyl)pyrrolo[3,2-b]pyridine-3-carboxamide | | Authors: | Chen, X.J, Lin, Q.M, Chen, Y.H. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.243 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of 5-Formyl-pyrrolo[3,2- b ]pyridine-3-carboxamides as New Selective, Potent, and Reversible-Covalent FGFR4 Inhibitors.
J.Med.Chem., 65, 2022
|
|
8QJR
 
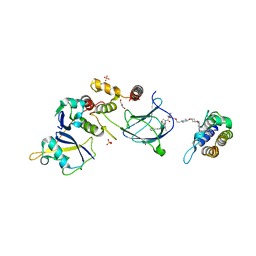 | | BRG1 bromodomain in complex with VBC via compound 17 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-[4-[3-[4-[(1R,5S)-3-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]-3,8-diazabicyclo[3.2.1]octan-8-yl]pyridin-2-yl]oxycyclobutyl]oxypiperidin-1-yl]ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJS
 
 | | VHL/Elongin B/Elongin C complex with compound 155 | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[2-(2-methoxyethoxy)ethoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[(1S)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.191 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
8QJT
 
 | | BRM (SMARCA2) Bromodomain in complex with ligand 10 | | Descriptor: | 2-[6-azanyl-5-[(1R,5S)-8-[2-(2-methoxyethoxy)pyridin-4-yl]-3,8-diazabicyclo[3.2.1]octan-3-yl]pyridazin-3-yl]phenol, CHLORIDE ION, Probable global transcription activator SNF2L2, ... | | Authors: | Kerry, P.S, Hole, A.J, Perez-Dorado, J.I. | | Deposit date: | 2023-09-13 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | PROTACs Targeting BRM (SMARCA2) Afford Selective In Vivo Degradation over BRG1 (SMARCA4) and Are Active in BRG1 Mutant Xenograft Tumor Models.
J.Med.Chem., 67, 2024
|
|
6L6Q
 
 | |
9B97
 
 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (1P)-N~3'~-[(2S)-3-cyclohexyl-1-(methylamino)-1-oxopropan-2-yl]-N~3~,N~3~-diethyl-4-fluoro-5'-{[4-(4-phenylbutyl)piperazin-1-yl]methyl}[1,1'-biphenyl]-3,3'-dicarboxamide, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B96
 
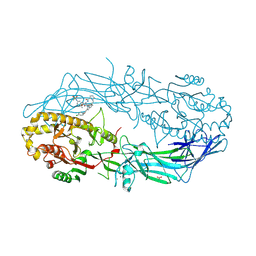 | | Crystal structure of the human PAD2 protein bound to inhibitor | | Descriptor: | 1-({(2P)-1-{(1R)-1-(2-bromophenyl)-3-[5-(methanesulfonamido)-2-methylanilino]-3-oxopropyl}-2-[3-(4-chlorophenoxy)phenyl]-1H-1,3-benzimidazol-6-yl}methyl)-N-methyl-D-prolinamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
9B98
 
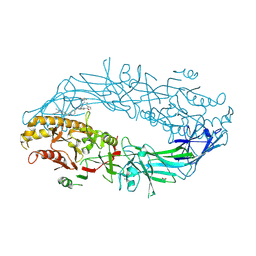 | | Crystal structure of the human PAD2 protein bound to small molecule | | Descriptor: | (5P)-N,N-diethyl-2-fluoro-5-(2-[({1-[2-(methylamino)-2-oxoethyl]cyclohexyl}methyl)amino]-6-{methyl[1-(2-methyl-1-phenyl-1H-1,3-benzimidazole-5-carbonyl)piperidin-4-yl]amino}pyrimidin-4-yl)benzamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Byrnes, L.J, Vajdos, F. | | Deposit date: | 2024-04-01 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Discovery, Characterization, and Structure of a Cell Active PAD2 Inhibitor Acting through a Novel Allosteric Mechanism.
Acs Chem.Biol., 19, 2024
|
|
6LBI
 
 | |
5WCD
 
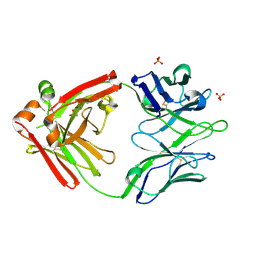 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 04-1D02 Fab. | | Descriptor: | PHOSPHATE ION, SULFATE ION, VRC315 04-1D02 Fab Heavy chain, ... | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-23 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.814 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
8Y7L
 
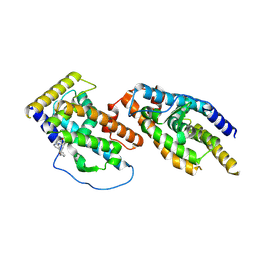 | |
5WCA
 
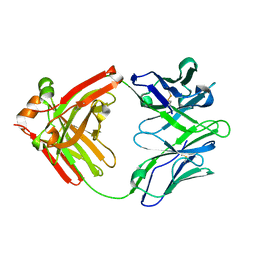 | | Crystal structure of the broadly neutralizing Influenza A antibody VRC 315 27-1C08 Fab. | | Descriptor: | VRC315 27-1C08 Fab Heavy chain, VRC315 27-1C08 Fab Light chain | | Authors: | Joyce, M.G, Andrews, S.F, Mascola, J.R, McDermott, A.B, Kwong, P.D. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans.
Sci Immunol, 2, 2017
|
|
6LBM
 
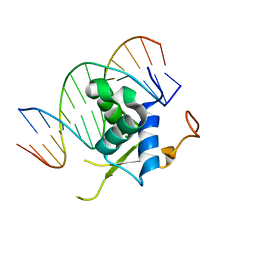 | |
2ATH
 
 | | Crystal structure of the ligand binding domain of human PPAR-gamma im complex with an agonist | | Descriptor: | 2-{5-[3-(7-PROPYL-3-TRIFLUOROMETHYLBENZO[D]ISOXAZOL-6-YLOXY)PROPOXY]INDOL-1-YL}ETHANOIC ACID, Peroxisome proliferator activated receptor gamma | | Authors: | Mahindroo, N, Huang, C.-F, Wu, S.-Y, Hsieh, H.-P. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Novel indole-based peroxisome proliferator-activated receptor agonists: design, SAR, structural biology, and biological activities
J.Med.Chem., 48, 2005
|
|
6EFC
 
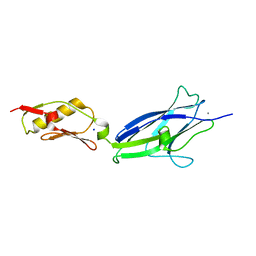 | | Hsa Siglec + Unique domains (unliganded) | | Descriptor: | CALCIUM ION, SODIUM ION, Streptococcal hemagglutinin | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6EFI
 
 | | SK678 binding region (Siglec + Unique) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Iverson, T.M. | | Deposit date: | 2018-08-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.715 Å) | | Cite: | Origins of glycan selectivity in streptococcal Siglec-like adhesins suggest mechanisms of receptor adaptation.
Nat Commun, 13, 2022
|
|
6L5N
 
 | | Crystal structure of human DEAD-box RNA helicase DDX21 at post-unwound state | | Descriptor: | MAGNESIUM ION, Nucleolar RNA helicase 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Chen, Z.J, Hu, X.J, Zhou, Z, Li, J.X. | | Deposit date: | 2019-10-24 | | Release date: | 2020-06-17 | | Last modified: | 2020-08-12 | | Method: | X-RAY DIFFRACTION (2.242 Å) | | Cite: | Structural Basis of Human Helicase DDX21 in RNA Binding, Unwinding, and Antiviral Signal Activation.
Adv Sci, 7, 2020
|
|
