1UEY
 
 | | Solution Structure of The First Fibronectin Type III Domain of Human KIAA0343 protein | | Descriptor: | KIAA0343 protein | | Authors: | Zhao, C, Kigawa, T, Muto, Y, Koshiba, S, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-22 | | Release date: | 2003-11-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The First Fibronectin Type III Domain of Human KIAA0343 protein
To be Published
|
|
1UG2
 
 | | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain | | Descriptor: | 2610100B20Rik gene product | | Authors: | Zhao, C, Kigawa, T, Tochio, N, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Mouse Hypothetical Gene (2610100B20Rik) Product Homologous to Myb DNA-binding Domain
To be Published
|
|
5D3E
 
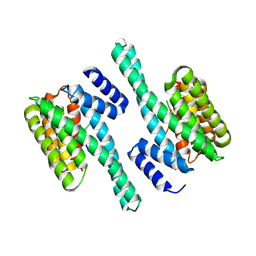 | |
3SM9
 
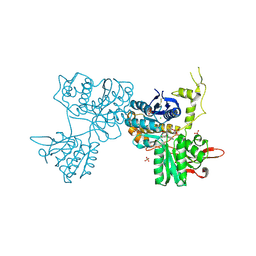 | | Crystal Structure of Metabotropic glutamate receptor 3 precursor in presence of LY341495 antagonist | | Descriptor: | 2-[(1S,2S)-2-carboxycyclopropyl]-3-(9H-xanthen-9-yl)-D-alanine, CHLORIDE ION, Metabotropic glutamate receptor 3, ... | | Authors: | Wernimont, A.K, Dong, A, Seitova, A, Crombet, L, Khutoreskaya, G, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Cossar, D, Dobrovetsky, E, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-27 | | Release date: | 2011-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure of Metabotropic glutamate receptor 3 precursor in presence of LY341495 antagonist
TO BE PUBLISHED
|
|
7R2S
 
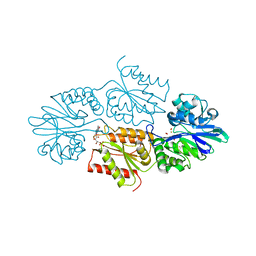 | | Crystal structure of a flavodiiron protein D52K mutant from Escherichia coli pressurized with krypton gas | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant from Escherichia coli pressurized with krypton gas
To Be Published
|
|
5D98
 
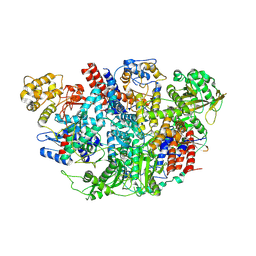 | | Influenza C Virus RNA-dependent RNA Polymerase - Space group P43212 | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Hengrung, N, El Omari, K, Serna Martin, I, Vreede, F.T, Cusack, S, Rambo, R.P, Vonrhein, C, Bricogne, G, Stuart, D.I, Grimes, J.M, Fodor, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the RNA-dependent RNA polymerase from influenza C virus.
Nature, 527, 2015
|
|
3SBO
 
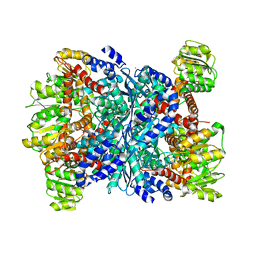 | | Structure of E.coli GDH from native source | | Descriptor: | CHLORIDE ION, NADP-specific glutamate dehydrogenase | | Authors: | Gee, C.L, Zubieta, C, Echols, N, Totir, M. | | Deposit date: | 2011-06-06 | | Release date: | 2012-03-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Macro-to-Micro Structural Proteomics: Native Source Proteins for High-Throughput Crystallization.
Plos One, 7, 2012
|
|
5DAY
 
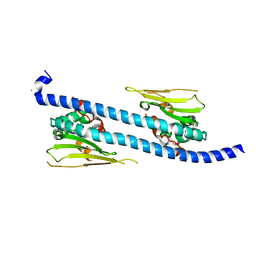 | | The structure of NAP1-Related Protein(NRP1) in Arabidopsis | | Descriptor: | CALCIUM ION, NAP1-related protein 1 | | Authors: | Zhu, Y, Rong, L, Yang, Y, Zhang, C, Feng, H.Y, Zheng, L.N, Shen, W.H, Ma, J.B, Dong, A.W. | | Deposit date: | 2015-08-20 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | The structure of NAP1-Related Protein(NRP1) in Arabidopsis
To Be Published
|
|
3S26
 
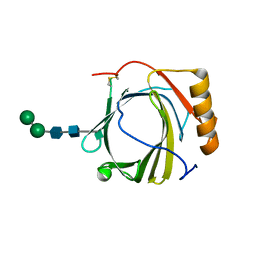 | | Crystal Structure of Murine Siderocalin (Lipocalin 2, 24p3) | | Descriptor: | Neutrophil gelatinase-associated lipocalin, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Correnti, C, Bandaranayake, A.D, Strong, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors.
Nucleic Acids Res., 39, 2011
|
|
5DCN
 
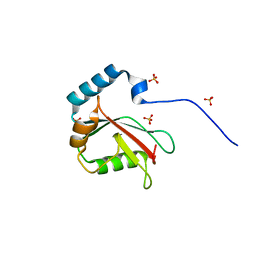 | | Crystal structure of LC3 in complex with TECPR2 LIR | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Stadel, D, Huber, J, Dotsch, V, Rogov, V.V, Behrends, C, Akutsu, M. | | Deposit date: | 2015-08-24 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | TECPR2 Cooperates with LC3C to Regulate COPII-Dependent ER Export.
Mol.Cell, 60, 2015
|
|
7R1J
 
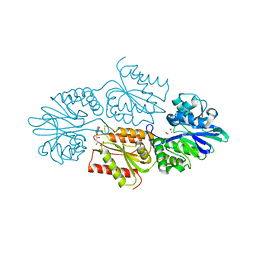 | | Crystal structure of a flavodiiron protein S262Y mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal structure of a flavodiiron protein S262Y mutant in the reduced state from Escherichia coli
To Be Published
|
|
7R2P
 
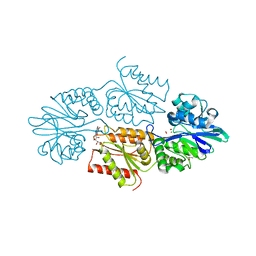 | | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.538 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the oxidized state from Escherichia coli
To Be Published
|
|
7R2X
 
 | |
7R0F
 
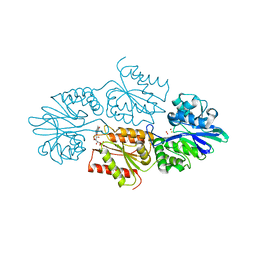 | | Crystal structure of a flavodiiron protein D52K mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-02 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant in the oxidized state from Escherichia coli
To Be Published
|
|
5DIE
 
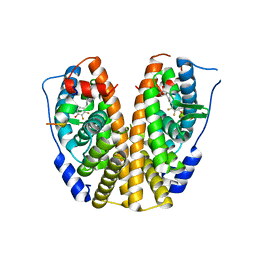 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a trifluoro-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-7a-methyl-5-(2,3,5-trifluoro-4-hydroxyphenyl)octahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-7a-methyl-5-(2,3,5-trifluoro-4-hydroxyphenyl)octahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7R2O
 
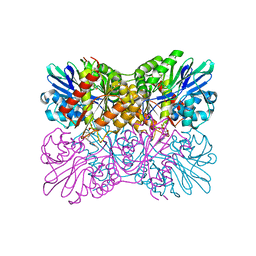 | | Crystal structure of a flavodiiron protein S262Y mutant in the oxidized state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a flavodiiron protein S262Y mutant in the oxidized state from Escherichia coli
To Be Published
|
|
5CUB
 
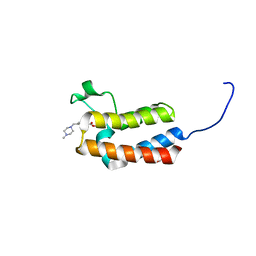 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 314268-40-1 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, methyl 4-[(4-methylpiperazin-1-yl)methyl]benzoate | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 314268-40-1 (SGC - Diamond I04-1 fragment screening)
To be published
|
|
7R2R
 
 | | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K/S262Y mutant in the reduced state from Escherichia coli
To Be Published
|
|
5CUG
 
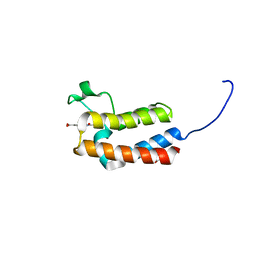 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 4-Acetamidobenzoic acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, PARA ACETAMIDO BENZOIC ACID | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 4-Acetamidobenzoic acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
7R1H
 
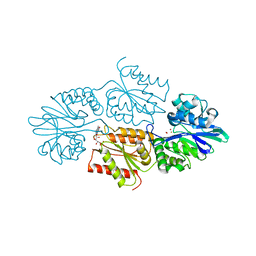 | | Crystal structure of a flavodiiron protein D52K mutant in the reduced state from Escherichia coli | | Descriptor: | Anaerobic nitric oxide reductase flavorubredoxin, FE (III) ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Borges, P.T, Teixeira, M, Romao, C.V, Frazao, C. | | Deposit date: | 2022-02-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a flavodiiron protein D52K mutant in the reduced state from Escherichia coli
To Be Published
|
|
5CRA
 
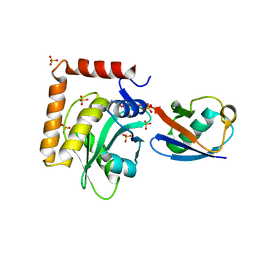 | | Structure of the SdeA DUB Domain | | Descriptor: | METHYL 4-AMINOBUTANOATE, Polyubiquitin-B, SULFATE ION, ... | | Authors: | Sheedlo, M.J, Qiu, J, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3SDJ
 
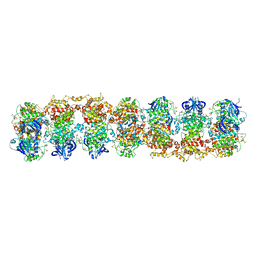 | | Structure of RNase-inactive point mutant of oligomeric kinase/RNase Ire1 | | Descriptor: | N~2~-1H-benzimidazol-5-yl-N~4~-(3-cyclopropyl-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | Authors: | Korennykh, A, Korostelev, A, Egea, P, Finer-Moore, J, Zhang, C, Stroud, R, Shokat, K, Walter, P. | | Deposit date: | 2011-06-09 | | Release date: | 2011-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural and functional basis for RNA cleavage by Ire1.
Bmc Biol., 9, 2011
|
|
3S0A
 
 | | Apis mellifera OBP14, native apo-protein | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
4CIK
 
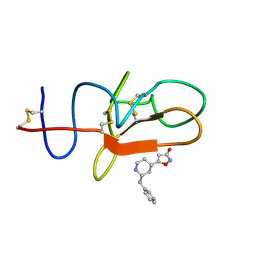 | | plasminogen kringle 1 in complex with inhibitor | | Descriptor: | 5-[(2R,4S)-2-(phenylmethyl)piperidin-4-yl]-1,2-oxazol-3-one, PLASMINOGEN | | Authors: | Xue, Y, Johansson, C, Cheng, L, Pettersen, D, Gustafsson, D. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of the Fibrinolysis Inhibitor Azd6564, Acting Via Interference of a Protein-Protein Interaction.
Acs Med.Chem.Lett., 5, 2014
|
|
3S0G
 
 | | Apis mellifera OBP 14 double mutant Gln44Cys, His97Cys | | Descriptor: | OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
