6QBJ
 
 | |
1SRY
 
 | |
6UWI
 
 | | Crystal structure of the Clostridium difficile translocase CDTb | | Descriptor: | ADP-ribosyltransferase binding component, CALCIUM ION | | Authors: | Pozharski, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the cell-binding component of theClostridium difficilebinary toxin reveals a di-heptamer macromolecular assembly.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UWO
 
 | |
6UWR
 
 | |
6UWT
 
 | |
6F5M
 
 | | Crystal structure of highly glycosylated human leukocyte elastase in complex with a thiazolidinedione inhibitor | | Descriptor: | 5-[[4-[[(2~{S})-4-methyl-1-oxidanylidene-1-[(2-propylphenyl)amino]pentan-2-yl]carbamoyl]phenyl]methyl]-2-oxidanylidene-1,3-thiazol-1-ium-4-olate, ACETATE ION, Neutrophil elastase, ... | | Authors: | Hochscherf, J, Pietsch, M, Tieu, W, Kuan, K, Hautmann, S, Abell, A, Guetschow, M, Niefind, K. | | Deposit date: | 2017-12-01 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of highly glycosylated human leukocyte elastase in complex with an S2' site binding inhibitor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1LYL
 
 | |
4QAG
 
 | | Structure of a dihydroxycoumarin active-site inhibitor in complex with the RNASE H domain of HIV-1 reverse transcriptase | | Descriptor: | (7,8-dihydroxy-2-oxo-2H-chromen-4-yl)acetic acid, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Himmel, D.M, Ho, W.C, Arnold, E. | | Deposit date: | 2014-05-04 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Structure of a Dihydroxycoumarin Active-Site Inhibitor in Complex with the RNase H Domain of HIV-1 Reverse Transcriptase and Structure-Activity Analysis of Inhibitor Analogs.
J.Mol.Biol., 426, 2014
|
|
4R5P
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | Descriptor: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-08-21 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2RDL
 
 | | Hamster Chymase 2 | | Descriptor: | Chymase 2, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYLKETONE INHIBITOR, SULFATE ION | | Authors: | Spurlino, J, Abad, M, Kervinen, J. | | Deposit date: | 2007-09-24 | | Release date: | 2007-10-30 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for elastolytic substrate specificity in rodent alpha-chymases.
J.Biol.Chem., 283, 2008
|
|
2RFX
 
 | | Crystal Structure of HLA-B*5701, presenting the self peptide, LSSPVTKSF | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Archbold, J.K, Macdonald, W.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-07-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human leukocyte antigen class I-restricted activation of CD8+ T cells provides the immunogenetic basis of a systemic drug hypersensitivity
Immunity, 28, 2008
|
|
4KRE
 
 | |
4K9F
 
 | | Neutron structure of Perdeuterated Rubredoxin refined against 1.75 resolution data collected on the new IMAGINE instrument at HFIR, ORNL | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-04-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | The IMAGINE instrument: first neutron protein structure and new capabilities for neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2RRN
 
 | | Solution structure of SecDF periplasmic domain P4 | | Descriptor: | Probable SecDF protein-export membrane protein | | Authors: | Tanaka, T, Tsukazaki, T, Echizen, Y, Nureki, O, Kohno, T. | | Deposit date: | 2011-01-30 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and function of a membrane component SecDF that enhances protein export
Nature, 474, 2011
|
|
4KJU
 
 | | Crystal structure of XIAP-Bir2 with a bound benzodiazepinone inhibitor. | | Descriptor: | E3 ubiquitin-protein ligase XIAP, N-{(3S)-5-(4-aminobenzoyl)-1-[(2-methoxynaphthalen-1-yl)methyl]-2-oxo-2,3,4,5-tetrahydro-1H-1,5-benzodiazepin-3-yl}-N~2~-methyl-L-alaninamide, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
4KRF
 
 | |
4KPP
 
 | | Crystal Structure of H+/Ca2+ Exchanger CAX | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, OLEIC ACID, ... | | Authors: | Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2013-05-14 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the counter-transport mechanism of a H+/Ca2+ exchanger.
Science, 341, 2013
|
|
4KJV
 
 | | Crystal structure of XIAP-Bir2 with a bound spirocyclic benzoxazepinone inhibitor. | | Descriptor: | 6-methoxy-5-({(3S)-3-[(N-methyl-L-alanyl)amino]-4-oxo-2',3,3',4,5',6'-hexahydro-5H-spiro[1,5-benzoxazepine-2,4'-pyran]-5-yl}methyl)naphthalene-2-carboxylic acid, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
8RIV
 
 | | T2R-TTL-1-K08 complex | | Descriptor: | (4-fluoranyl-2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E, Prota, A.E.P. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8RIW
 
 | | T2R-TTL-1-L01 complex | | Descriptor: | (2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E.P, Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E. | | Deposit date: | 2023-12-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8RPO
 
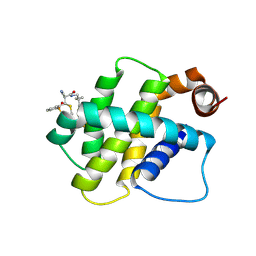 | | BFL1 in complex with a reversible covalent ligand | | Descriptor: | (~{E})-2-cyano-3-(2-dimethylphosphorylphenyl)-~{N}-[[1-[4-(trifluoromethyl)phenyl]cyclopropyl]methyl]prop-2-enamide, Bcl-2-related protein A1 | | Authors: | Hargreaves, D. | | Deposit date: | 2024-01-16 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Identification and Evaluation of Reversible Covalent Binders to Cys55 of Bfl-1 from a DNA-Encoded Chemical Library Screen.
Acs Med.Chem.Lett., 15, 2024
|
|
8RC6
 
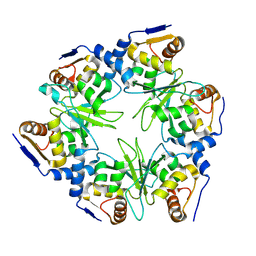 | |
4UYD
 
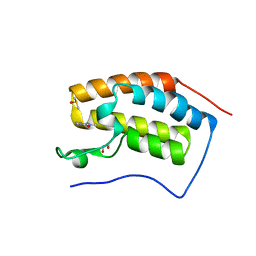 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 1,3-dimethyl-2-oxo-2,3- dihydro-1H-1,3-benzodiazole-5-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxamide, BROMODOMAIN-CONTAINING PROTEIN 4, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2014-08-30 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | 1,3-Dimethyl Benzimidazolones are Potent, Selective Inhibitors of the Brpf1 Bromodomain.
Acs Med.Chem.Lett., 5, 2014
|
|
4UYE
 
 | | BROMODOMAIN OF HUMAN BRPF1 WITH N-1,3-dimethyl-2-oxo-6-(piperidin-1- yl)-2,3-dihydro-1H-1,3-benzodiazol-5-yl-2-methoxybenzamide | | Descriptor: | 1,2-ETHANEDIOL, N-[1,3-dimethyl-2-oxo-6-(piperidin-1-yl)-2,3-dihydro-1H-benzimidazol-5-yl]-2-methoxybenzamide, PEREGRIN | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2014-08-30 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 1,3-Dimethyl Benzimidazolones are Potent, Selective Inhibitors of the Brpf1 Bromodomain.
Acs Med.Chem.Lett., 5, 2014
|
|
