8X2Z
 
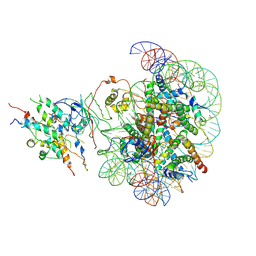 | | The class2 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X32
 
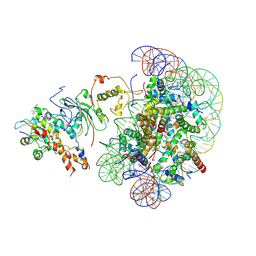 | | The piccolo NuA4 bound to the H2A.Z nucleosome-H4KQ Complex with Ac-CoA at resetting state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X30
 
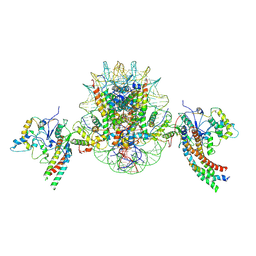 | | Structure of piccolo NuA4 and H2A.Z nucleosome 2:1 complex | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2Y
 
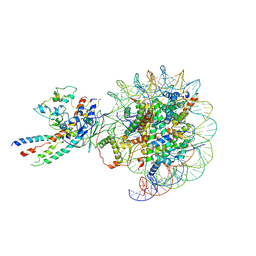 | | The class1 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2X
 
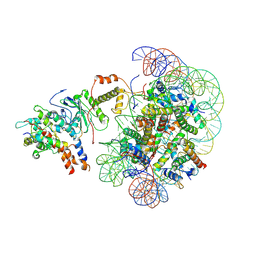 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex at pre-H4-acetylation state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X31
 
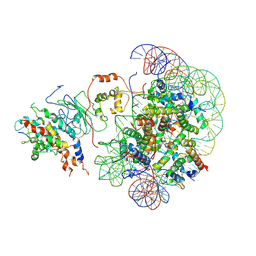 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex with Ac-CoA at resetting state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
7V67
 
 | |
7VRD
 
 | |
7W8S
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Su, C, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular Basis of Mink ACE2 Binding to SARS-CoV-2 and Its Mink-Derived Variants.
J.Virol., 96, 2022
|
|
7WA1
 
 | |
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
9ERS
 
 | | Hydrogenase-2 Ni-C state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Wong, K.L, Carr, S.B, Ash, P.A, Vincent, K.A. | | Deposit date: | 2024-03-25 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Glutamate flick enables proton tunnelling during fast redox biocatalysis
To Be Published
|
|
5UFU
 
 | | Structure of AMPK bound to activator | | Descriptor: | 1,4:3,6-dianhydro-2-O-(6-chloro-5-{4-[1-(hydroxymethyl)cyclopropyl]phenyl}-1H-benzimidazol-2-yl)-D-mannitol, 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2017-01-05 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Activation of Skeletal Muscle AMPK Promotes Glucose Disposal and Glucose Lowering in Non-human Primates and Mice.
Cell Metab., 25, 2017
|
|
9ERA
 
 | | Hydrogenase-1 Ni-Lii state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Wong, K.L, Carr, S.B, Ash, P.A, Vincent, K.A. | | Deposit date: | 2024-03-22 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Glutamate flick enables proton tunnelling during fast redox biocatalysis
To Be Published
|
|
5VAG
 
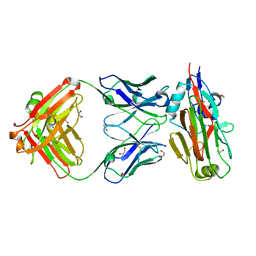 | | Crystal structure of H7-specific antibody m826 in complex with the HA1 domain of hemagglutinin from H7N9 influenza virus | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antibody m826, Hemagglutinin, ... | | Authors: | Song, H, Ying, T, Ji, X. | | Deposit date: | 2017-03-26 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Potent Germline-like Human Monoclonal Antibody Targets a pH-Sensitive Epitope on H7N9 Influenza Hemagglutinin.
Cell Host Microbe, 22, 2017
|
|
5VLK
 
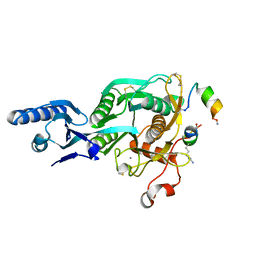 | |
5VLP
 
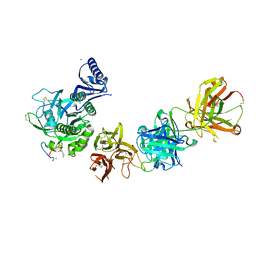 | | PCSK9 complex with LDLR antagonist peptide and Fab7G7 | | Descriptor: | Fab7G7 heavy chain, Fab7G7 light chain, LDLR antagonist peptide, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VL7
 
 | | PCSK9 complex with Fab33 | | Descriptor: | Fab33 heavy chain, Fab33 light chain, Proprotein convertase subtilisin/kexin type 9 | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLA
 
 | | Short PCSK9 delta-P' complex with Fusion2 peptide | | Descriptor: | CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-MET-PRO-TRP-ASN-LEU-VAL-ARG-ILE-GLY-LEU-LEU | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7WLP
 
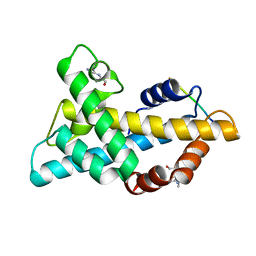 | |
5VLH
 
 | | Short PCSK9 delta-P' complex with peptide Pep1 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-ARG-LEU-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-PRO-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5VLL
 
 | | Short PCSK9 delta-P' complex with peptide Pep3 | | Descriptor: | ACE-THR-VAL-PHE-THR-SER-TRP-GLU-GLU-TYR-LEU-ASP-TRP-VAL-NH2, CALCIUM ION, CYS-PHE-ILE-PRO-TRP-ASN-LEU-GLN-ARG-ILE-GLY-LEU-LEU-CYS, ... | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2017-04-25 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Discovery of a cryptic peptide-binding site on PCSK9 and design of antagonists.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7EKE
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKG
 
 | | Structure of SARS-CoV-2 Beta variant spike receptor-binding domain complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EKH
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Han, P.C, Su, C, Zhang, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2021-04-05 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
