5USG
 
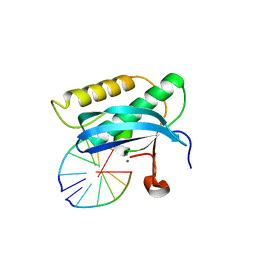 | | 5-Se-T2/4-DNA and native RNA hybrid in complex with RNase H catalytic domain D132N mutant | | Descriptor: | DNA (5'-D(*AP*(T5S)P*GP*(T5S)P*CP*G)-3'), MAGNESIUM ION, RNA (5'-R(*UP*CP*GP*AP*CP*A)-3'), ... | | Authors: | Fang, Z, Qin, L, Huang, Z. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
5VBU
 
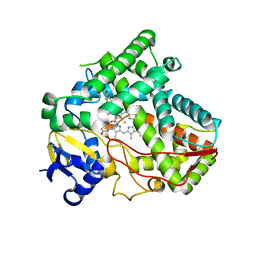 | |
2HMI
 
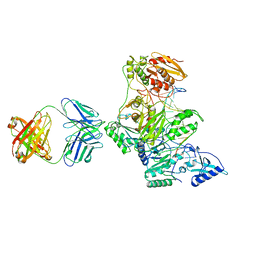 | | HIV-1 REVERSE TRANSCRIPTASE/FRAGMENT OF FAB 28/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3'), FAB FRAGMENT OF MONOCLONAL ANTIBODY 28, ... | | Authors: | Ding, J, Arnold, E. | | Deposit date: | 1998-04-10 | | Release date: | 1998-10-14 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody Fab fragment at 2.8 A resolution.
J.Mol.Biol., 284, 1998
|
|
7V4Y
 
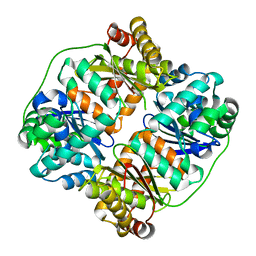 | | TTHA1264/TTHA1265 complex | | Descriptor: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | Authors: | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | Deposit date: | 2021-08-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
2MQW
 
 | |
2K8Y
 
 | | Solution NMR Structure of Cgi121 from Methanococcus jannaschii. Northeast Structural Genomics Consortium Target MJ0187 | | Descriptor: | Uncharacterized protein MJ0187 | | Authors: | Rumpel, S, Fares, C, Neculai, D, Arrowsmith, C, Montelione, G.T, Sicheri, F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-25 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine.
Mol.Cell, 32, 2008
|
|
2MRI
 
 | |
2MR3
 
 | | A subunit of 26S proteasome lid complex | | Descriptor: | 26S proteasome regulatory subunit RPN9 | | Authors: | Wu, Y, Hu, Y, Jin, C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-02-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of yeast Rpn9: insights into proteasome lid assembly.
J. Biol. Chem., 290, 2015
|
|
3HLC
 
 | | Simvastatin Synthase (LovD) from Aspergillus terreus, S5 mutant, unliganded | | Descriptor: | GLYCEROL, TETRAETHYLENE GLYCOL, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Laidman, J, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLD
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant complex with monacolin J acid | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,6R,8S,8aR)-8-hydroxy-2,6-dimethyl-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]heptanoic acid, FORMIC ACID, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLG
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant, S76A mutant, complex with lovastatin | | Descriptor: | (3R,5R)-7-((1R,2R,6S,8R,8AS)-2,6-DIMETHYL-8-{[(2R)-2-METHYLBUTANOYL]OXY}-1,2,6,7,8,8A-HEXAHYDRONAPHTHALEN-1-YL)-3,5-DIHYDROXYHEPTANOIC ACID, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLE
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant, S76A mutant, complex with monacolin J acid | | Descriptor: | (3R,5R)-3,5-dihydroxy-7-[(1S,2S,6R,8S,8aR)-8-hydroxy-2,6-dimethyl-1,2,6,7,8,8a-hexahydronaphthalen-1-yl]heptanoic acid, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLF
 
 | | Simvastatin Synthase (LovD), from Aspergillus terreus, S5 mutant, S76A mutant, complex with simvastatin | | Descriptor: | Simvastatin acid, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HL9
 
 | | Simvastatin Synthase (LovD) from Aspergillus terreus, unliganded | | Descriptor: | Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Laidman, J, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
3HLB
 
 | | Simvastatin Synthase (LovD) from Aspergillus terreus, unliganded, selenomethionyl derivative | | Descriptor: | SULFATE ION, Transesterase | | Authors: | Sawaya, M.R, Yeates, T.O, Laidman, J, Pashkov, I, Gao, X, Tang, Y. | | Deposit date: | 2009-05-27 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Directed evolution and structural characterization of a simvastatin synthase
Chem.Biol., 16, 2009
|
|
7MET
 
 | | A. baumannii MsbA in complex with TBT1 decoupler | | Descriptor: | 2-(4-chlorobenzamido)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxylic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
7MEW
 
 | | E. coli MsbA in complex with G247 | | Descriptor: | (2E)-3-{1-cyclopropyl-7-[(1S)-1-(3,6-dichloro-2-fluorophenyl)ethoxy]naphthalen-2-yl}prop-2-enoic acid, ATP-dependent lipid A-core flippase | | Authors: | Thelot, F, Liao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Distinct allosteric mechanisms of first-generation MsbA inhibitors.
Science, 374, 2021
|
|
8F2M
 
 | |
3IK9
 
 | | Human GST A1-1-GIMF with GSDHN | | Descriptor: | (S)-2-amino-5-((R)-1-(carboxymethylamino)-3-((3S,4R)-1,4-dihydroxynonan-3-ylthio)-1-oxopropan-2-ylamino)-5-oxopentanoic acid, Glutathione S-transferase A1 | | Authors: | Balogh, L.M, Le Trong, I, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal.
Biochemistry, 49, 2010
|
|
3IK7
 
 | | Human glutathione transferase a4-4 with GSDHN | | Descriptor: | (S)-2-amino-5-((R)-1-(carboxymethylamino)-3-((3S,4R)-1,4-dihydroxynonan-3-ylthio)-1-oxopropan-2-ylamino)-5-oxopentanoic acid, Glutathione S-transferase A4, SULFATE ION | | Authors: | Balogh, L.M, Le Trong, I, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2009-08-05 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Substrate specificity combined with stereopromiscuity in glutathione transferase A4-4-dependent metabolism of 4-hydroxynonenal.
Biochemistry, 49, 2010
|
|
8GRY
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with rat ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-03 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
6NAK
 
 | | BACTERIAL PROTEIN COMPLEX TM BDE complex | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, TsaE, ... | | Authors: | Stec, B. | | Deposit date: | 2018-12-05 | | Release date: | 2023-04-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Discovery of the Universal tRNA Binding Mode for the TsaD-like Components of the t6A tRNA Modification Pathway
Biophysica, 3, 2023
|
|
7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
8H06
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8H5C
 
 | | Structure of SARS-CoV-2 Omicron BA.2.75 RBD in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Bai, B, Liu, K.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-10-12 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
