8I6K
 
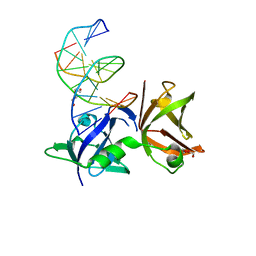 | | Structure of hMNDA HIN with dsDNA | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DNA (5'-D(*GP*GP*CP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Li, Y.L, Jin, T.C. | | Deposit date: | 2023-01-28 | | Release date: | 2023-07-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of dsDNA recognition by the hMNDA HIN domain: New insights into the DNA-binding model of a PYHIN protein.
Int.J.Biol.Macromol., 245, 2023
|
|
6JPE
 
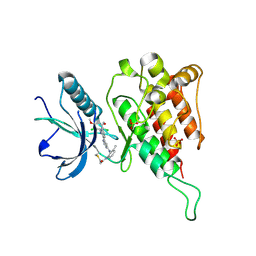 | | Crystal structure of FGFR4 kinase domain with irreversible inhibitor 1 | | Descriptor: | Fibroblast growth factor receptor 4, N-[2-[[6-[2-[[2,6-bis(chloranyl)-3,5-dimethoxy-phenyl]amino]pyridin-3-yl]pyrimidin-4-yl]amino]-3-methyl-phenyl]prop-2-enamide, SULFATE ION | | Authors: | Chen, X, Dai, S, Zhou, Z, Chen, Y. | | Deposit date: | 2019-03-26 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Development of a Potent and Specific FGFR4 Inhibitor for the Treatment of Hepatocellular Carcinoma.
J.Med.Chem., 63, 2020
|
|
7YG7
 
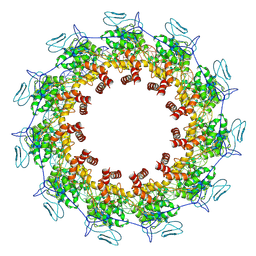 | | Structure of the Spring Viraemia of Carp Virus ribonucleoprotein Complex | | Descriptor: | Nucleoprotein, RNA (99-mer) | | Authors: | Liu, B, Wang, Z.X, Yang, T, Yu, D.Q, Ouyang, Q. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-15 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the Spring Viraemia of Carp Virus Ribonucleoprotein Complex Reveals Its Assembly Mechanism and Application in Antiviral Drug Screening.
J.Virol., 97, 2023
|
|
8IUO
 
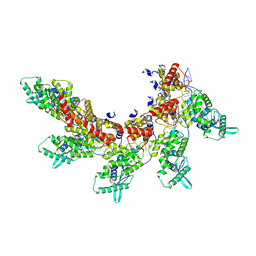 | | respiratory syncytial virus nucleocapsid-like assembly | | Descriptor: | Nucleoprotein, RNA (35-MER) | | Authors: | Wang, Y, Luo, Y, Ling, X, Luo, B, Jia, G, Dong, H, Su, Z. | | Deposit date: | 2023-03-24 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of the nucleocapsid-like assembly of respiratory syncytial virus.
Signal Transduct Target Ther, 8, 2023
|
|
8Y6U
 
 | |
5CM4
 
 | | Crystal structure of human Frizzled 4 Cysteine-Rich Domain (CRD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4 | | Authors: | Ke, J, Parker, N, Gu, X, Zhang, C, Melcher, K, Xu, H.E. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
6UJG
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-{5-[(3S)-3-{[5-(acetylamino)-1,3,4-thiadiazol-2-yl]amino}pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6UMD
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-(pyridin-3-yl)-N-(5-{4-[(5-{[(pyridin-3-yl)acetyl]amino}-1,3,4-thiadiazol-2-yl)amino]piperidin-1-yl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6UME
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-(pyridin-3-yl)-N-(5-{4-[(5-{[(pyridin-3-yl)acetyl]amino}-1,3,4-thiadiazol-2-yl)amino]piperidin-1-yl}-1,3,4-thiadiazol-2-yl)acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6UMC
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-phenyl-N-{5-[(3R)-3-({5-[(phenylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
6UMF
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N-(5-{[1-(5-amino-1,3,4-thiadiazol-2-yl)piperidin-4-yl]oxy}-1,3,4-thiadiazol-2-yl)-2-phenylacetamide | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-09 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00012
To Be Published
|
|
7JTQ
 
 | |
7JTN
 
 | |
7WK5
 
 | | Cryo-EM structure of Omicron S-ACE2, C2 state | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Han, W.Y, Wang, Y.F. | | Deposit date: | 2022-01-08 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant receptor engagement and antibody evasion and neutralization
Biorxiv, 2022
|
|
7WK3
 
 | | SARS-CoV-2 Omicron S-open | | Descriptor: | Spike glycoprotein | | Authors: | Li, J.W, Cong, Y. | | Deposit date: | 2022-01-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for ACE2 engagement and antibody evasion and neutralization of SARS-Co-2 Omicron varient
To Be Published
|
|
7WLK
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with Otilonium Bromide(OB) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-[diethyl(methyl)-$l^{4}-azanyl]ethyl 4-[(2-octoxyphenyl)carbonylamino]benzoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLL
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with pimozide(PMZ) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[1-[4,4-bis(4-fluorophenyl)butyl]piperidin-4-yl]-1~{H}-benzimidazol-2-one, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7WLJ
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with mibefradil (MIB) | | Descriptor: | (1S,2S)-2-(2-{[3-(1H-benzimidazol-2-yl)propyl](methyl)amino}ethyl)-6-fluoro-1-(propan-2-yl)-1,2,3,4-tetrahydronaphthalen-2-yl methoxyacetate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
7C91
 
 | | Blasnase-T13A with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7CBU
 
 | | Blasnase-T13A with L-Asp | | Descriptor: | ASPARTIC ACID, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7C8Q
 
 | | Blasnase-T13A with D-asn | | Descriptor: | Asparaginase, D-ASPARAGINE, FORMIC ACID, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7CBW
 
 | | Blasnase-T13A with D-asn | | Descriptor: | FORMIC ACID, L-asparaginase, MAGNESIUM ION | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-15 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7CB4
 
 | | Crystal structures of of BlAsnase | | Descriptor: | FORMIC ACID, GLYCEROL, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7C8X
 
 | | Blasnase-T13A with L-asn | | Descriptor: | ASPARAGINE, Asparaginase, FORMIC ACID, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
7CBR
 
 | | Blasnase-T13A with D-asn | | Descriptor: | D-ASPARAGINE, FORMIC ACID, L-asparaginase, ... | | Authors: | Lu, F, Ran, T, Jiao, L, Wang, W. | | Deposit date: | 2020-06-13 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of l-asparaginase from Bacillus licheniformis Reveal an Essential Residue for its Substrate Stereoselectivity.
J.Agric.Food Chem., 69, 2021
|
|
