4DRQ
 
 | | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52: Complex of FKBP51 with 2-(3-((R)-1-((S)-1-(3,5-dichlorophenylsulfonyl)piperidine-2-carbonyloxy)-3-(3,4-dimethoxy -phenyl)propyl)phenoxy)acetic acid | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP5, {3-[(1S)-1-[({(2S)-1-[(3,5-dichlorophenyl)sulfonyl]piperidin-2-yl}carbonyl)oxy]-3-(3,4-dimethoxyphenyl)propyl]phenoxy}acetic acid | | Authors: | Gopalakrishnan, R, Kozany, C, Wang, Y, Hoogeland, B, Bracher, A, Hausch, F, Schneider, S. | | Deposit date: | 2012-02-17 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Exploration of Pipecolate Sulfonamides as Binders of the FK506-Binding Proteins 51 and 52.
J.Med.Chem., 55, 2012
|
|
7V6N
 
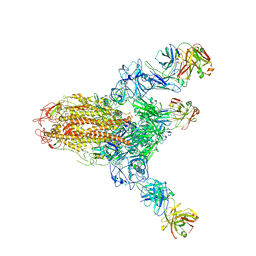 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1 | | Descriptor: | 111 H, 111 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-20 | | Release date: | 2022-09-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 111 state1
to be published
|
|
7V6O
 
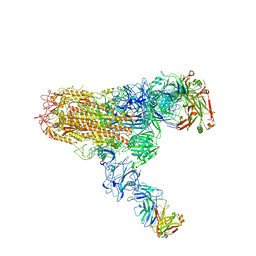 | | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2) | | Descriptor: | 111 H, 111 L, Spike glycoprotein | | Authors: | Wang, X, Zhao, J, Wang, Z, Zeng, J, Zhang, S, Wang, Y. | | Deposit date: | 2021-08-20 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.56 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 111 (state 2)
to be published
|
|
4EYR
 
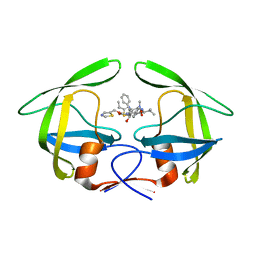 | | Crystal structure of multidrug-resistant clinical isolate 769 HIV-1 protease in complex with ritonavir | | Descriptor: | HIV-1 PROTEASE, RITONAVIR | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Brunzelle, J.S, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-05-01 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the mechanism of drug resistance: X-ray structure analysis of multi-drug resistant HIV-1 protease ritonavir complex.
Biochem.Biophys.Res.Commun., 431, 2013
|
|
7V3L
 
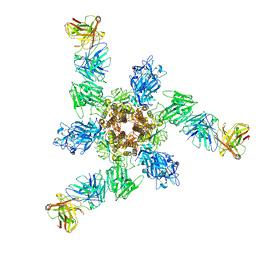 | | MERS S ectodomain trimer in complex with neutralizing antibody 6516 | | Descriptor: | Spike glycoprotein, antibody H, antibody L | | Authors: | Wang, X, Zhao, J, Wang, Z, Wang, Y, Zeng, J. | | Deposit date: | 2021-08-10 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | MERS S ectodomain trimer in complex with neutralizing antibody 6516
to be published
|
|
4GYE
 
 | | MDR 769 HIV-1 Protease in Complex with Reduced P1F | | Descriptor: | P1F peptide, Protease | | Authors: | Dewdney, T.G, Wang, Y, Brunzelle, J, Reiter, S.J, Kovari, I.A, Kovari, L.C. | | Deposit date: | 2012-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
3CIG
 
 | | Crystal structure of mouse TLR3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
4EKU
 
 | |
3CPU
 
 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | Deposit date: | 1999-06-08 | | Release date: | 2001-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
4P5K
 
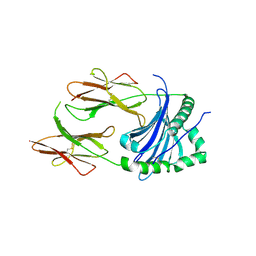 | | Structural Basis of Chronic Beryllium Disease: Bridging the Gap Between Allergy and Autoimmunity | | Descriptor: | HLA class II histocompatibility antigen, DP alpha 1 chain, RAS peptide,HLA class II histocompatibility antigen, ... | | Authors: | Clayton, G.M, Wang, Y, Crawford, F, Novikov, A, Wimberly, B.T, Kieft, J.S, Falta, M.T, Bowerman, N.A, Marrack, P, Fontenot, A.P, Dai, S, Kappler, J.W. | | Deposit date: | 2014-03-17 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis of chronic beryllium disease: linking allergic hypersensitivity and autoimmunity.
Cell, 158, 2014
|
|
4GZF
 
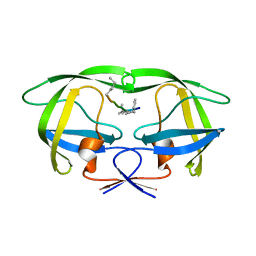 | | Multi-drug resistant HIV-1 protease 769 variant with reduced LrF peptide | | Descriptor: | LrF peptide, Protease | | Authors: | Dewdney, T.G, Wang, Y, Kovari, I.A, Brunzelle, J.S, Reiter, S.J, Kovari, L.C. | | Deposit date: | 2012-09-06 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ligand modifications to reduce the relative resistance of multi-drug resistant HIV-1 protease.
Bioorg.Med.Chem., 21, 2013
|
|
4RXA
 
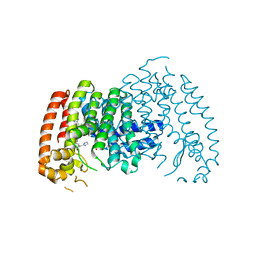 | | Crystal structure of human farnesyl diphosphate synthase in complex with BPH-1358 | | Descriptor: | Farnesyl pyrophosphate synthase, N,N'-bis[3-(4,5-dihydro-1H-imidazol-2-yl)phenyl]biphenyl-4,4'-dicarboxamide, PHOSPHATE ION | | Authors: | Liu, Y.-L, Cao, R, Wang, Y, Oldfield, E. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Farnesyl diphosphate synthase inhibitors with unique ligand-binding geometries.
ACS Med Chem Lett, 6, 2015
|
|
8JFU
 
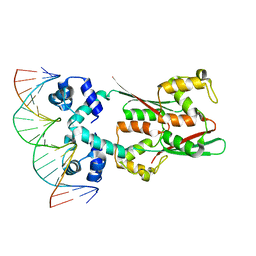 | | AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFO
 
 | |
8JFR
 
 | | N-terminal domain of AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JG9
 
 | |
8JFT
 
 | | Cryo-EM structure of SaCas9-AcrIIA15 CTD-sgRNA complex | | Descriptor: | AcrIIA15, CRISPR-associated endonuclease Cas9, sgRNA of SaCas9 | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
6AVS
 
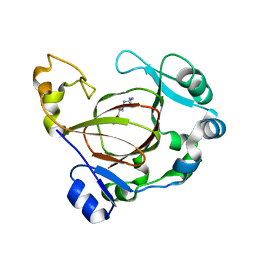 | | Complex structure of JMJD5 and Symmetric Monomethyl-Arginine (MMA) | | Descriptor: | (2S)-2-amino-5-[(N-methylcarbamimidoyl)amino]pentanoic acid, Lysine-specific demethylase 8, ZINC ION | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-04 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
6AX3
 
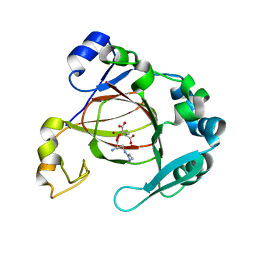 | | Complex structure of JMJD5 and Symmetric Dimethyl-Arginine (SDMA) | | Descriptor: | 2-OXOGLUTARIC ACID, Lysine-specific demethylase 8, N3, ... | | Authors: | Lee, S, Liu, H, Wang, Y, Dai, S, Zhang, G. | | Deposit date: | 2017-09-06 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Specific Recognition of Arginine Methylated Histone Tails by JMJD5 and JMJD7.
Sci Rep, 8, 2018
|
|
4KO5
 
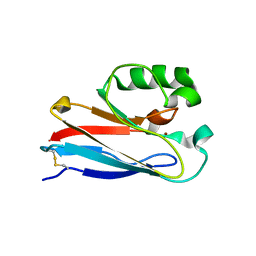 | | Investigating the functional significance of the interlocked pair structural determinants in Pseudomonas aeruginosa azurin (V31I/W48L/V95I/Y108F) | | Descriptor: | Azurin, COPPER (II) ION | | Authors: | Inampudi, K.K, Wang, Y, Meng, W, Tobin, P.H, Wilson, C.J. | | Deposit date: | 2013-05-11 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Investigating the functional significance of the interlocked pair structural determinants in Pseudomonas aeruginosa azurin (V31I/W48L/V95I/Y108F)
To be Published
|
|
6KH3
 
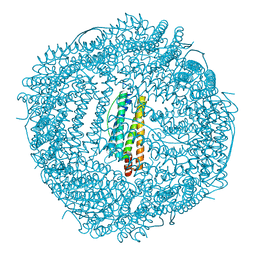 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
6KH4
 
 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
6KH5
 
 | | Design and crystal structure of protein MOFs with ferritin nanocages as linkers and nickel clusters as nodes | | Descriptor: | FE (III) ION, Ferritin, NICKEL (II) ION | | Authors: | Gu, C, Chen, H, Wang, Y, Zhang, T, Wang, H, Zhao, G. | | Deposit date: | 2019-07-12 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Structural Insight into Binary Protein Metal-Organic Frameworks with Ferritin Nanocages as Linkers and Nickel Clusters as Nodes.
Chemistry, 26, 2020
|
|
4L1A
 
 | | Crystallographic study of multi-drug resistant HIV-1 protease Lopinavir complex: mechanism of drug recognition and resistance | | Descriptor: | MDR769 HIV-1 protease, N-{1-BENZYL-4-[2-(2,6-DIMETHYL-PHENOXY)-ACETYLAMINO]-3-HYDROXY-5-PHENYL-PENTYL}-3-METHYL-2-(2-OXO-TETRAHYDRO-PYRIMIDIN-1-YL)-BUTYRAMIDE | | Authors: | Liu, Z, Yedidi, R.S, Wang, Y, Dewdney, T, Reiter, S, Brunzelle, J, Kovari, I, Kovari, L. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic study of multi-drug resistant HIV-1 protease lopinavir complex: mechanism of drug recognition and resistance.
Biochem.Biophys.Res.Commun., 437, 2013
|
|
4KVP
 
 | | Human p53 Core Domain Mutant V157F | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
