2M56
 
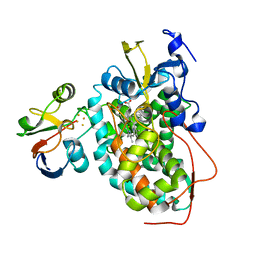 | | The structure of the complex of cytochrome P450cam and its electron donor putidaredoxin determined by paramagnetic NMR spectroscopy | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hiruma, Y, Hass, M.A.S, Ubbink, M. | | Deposit date: | 2013-02-14 | | Release date: | 2013-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography.
J.Mol.Biol., 425, 2013
|
|
2ME2
 
 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2LXL
 
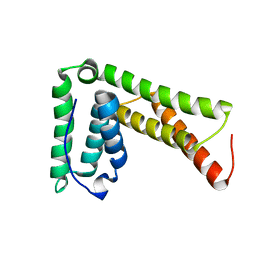 | | Lip5(mit)2 | | Descriptor: | Vacuolar protein sorting-associated protein VTA1 homolog | | Authors: | Skalicky, J.J, Sundquist, W.I. | | Deposit date: | 2012-08-29 | | Release date: | 2012-11-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Interactions of the Human LIP5 Regulatory Protein with Endosomal Sorting Complexes Required for Transport.
J.Biol.Chem., 287, 2012
|
|
2K8A
 
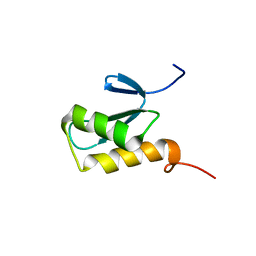 | | Solution structure of a novel Ubiquitin-binding domain from Human PLAA (PFUC, Gly76-Pro77 trans isomer) | | Descriptor: | Phospholipase A-2-activating protein | | Authors: | Fu, Q.S, Zhou, C.J, Gao, H.C, Lin, D.H, Hu, H.Y. | | Deposit date: | 2008-09-04 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Ubiquitin Recognition by a Novel Domain from Human Phospholipase A2-activating Protein.
J.Biol.Chem., 284, 2009
|
|
2KVA
 
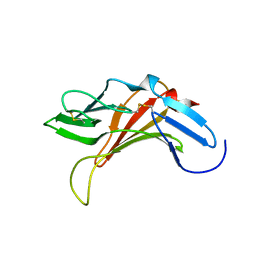 | |
1AYM
 
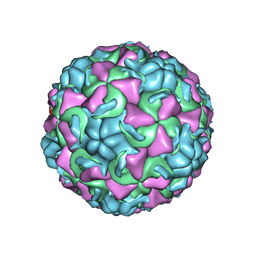 | | HUMAN RHINOVIRUS 16 COAT PROTEIN AT HIGH RESOLUTION | | Descriptor: | HUMAN RHINOVIRUS 16 COAT PROTEIN, LAURIC ACID, MYRISTIC ACID, ... | | Authors: | Hadfield, A.T, Rossmann, M.G. | | Deposit date: | 1997-11-06 | | Release date: | 1998-01-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The refined structure of human rhinovirus 16 at 2.15 A resolution: implications for the viral life cycle.
Structure, 5, 1997
|
|
1BJR
 
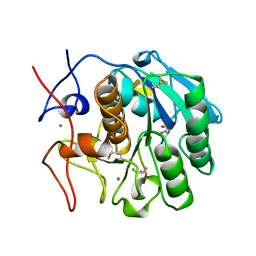 | | COMPLEX FORMED BETWEEN PROTEOLYTICALLY GENERATED LACTOFERRIN FRAGMENT AND PROTEINASE K | | Descriptor: | CALCIUM ION, LACTOFERRIN, PROTEINASE K | | Authors: | Singh, T.P, Sharma, S, Karthikeyan, S, Betzel, C, Bhatia, K.L. | | Deposit date: | 1998-06-27 | | Release date: | 1998-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Crystal structure of a complex formed between proteolytically-generated lactoferrin fragment and proteinase K.
Proteins, 33, 1998
|
|
1BTU
 
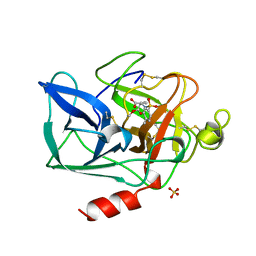 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3S, 4R)-1-TOLUENESULPHONYL-3-ETHYL-AZETIDIN-2-ONE-4-CARBOXYLIC ACID | | Descriptor: | (3R)-3-ethyl-N-[(4-methylphenyl)sulfonyl]-L-aspartic acid, CALCIUM ION, ELASTASE, ... | | Authors: | Wilmouth, R.C, Clifton, I.J, Schofield, C.J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of elastase by N-sulfonylaryl beta-lactams: anatomy of a stable acyl-enzyme complex.
Biochemistry, 37, 1998
|
|
1E2A
 
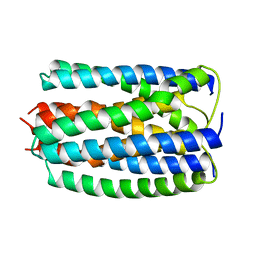 | | ENZYME IIA FROM THE LACTOSE SPECIFIC PTS FROM LACTOCOCCUS LACTIS | | Descriptor: | ENZYME IIA, MAGNESIUM ION | | Authors: | Sliz, P, Engelmann, R, Hengstenberg, W, Pai, E.F. | | Deposit date: | 1997-04-25 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of enzyme IIAlactose from Lactococcus lactis reveals a new fold and points to possible interactions of a multicomponent system.
Structure, 5, 1997
|
|
1CQH
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN REF-1 (RESIDUES 59-71 OF THE P50 SUBUNIT OF NFKB), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | REF-1 PEPTIDE, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1996-04-02 | | Release date: | 1996-08-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal.
Structure, 4, 1996
|
|
1CQG
 
 | | HIGH RESOLUTION SOLUTION NMR STRUCTURE OF MIXED DISULFIDE INTERMEDIATE BETWEEN HUMAN THIOREDOXIN (C35A, C62A, C69A, C73A) MUTANT AND A 13 RESIDUE PEPTIDE COMPRISING ITS TARGET SITE IN HUMAN REF-1 (RESIDUES 59-71 OF THE P50 SUBUNIT OF NFKB), NMR, 31 STRUCTURES | | Descriptor: | REF-1 PEPTIDE, THIOREDOXIN | | Authors: | Clore, G.M, Qin, J, Gronenborn, A.M. | | Deposit date: | 1996-04-02 | | Release date: | 1996-08-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal.
Structure, 4, 1996
|
|
1D3E
 
 | |
1FKA
 
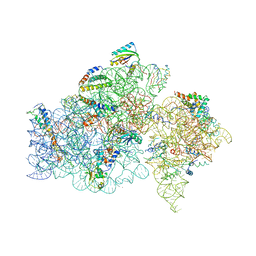 | | STRUCTURE OF FUNCTIONALLY ACTIVATED SMALL RIBOSOMAL SUBUNIT AT 3.3 A RESOLUTION | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Schluenzen, F, Tocilj, A, Zarivach, R, Harms, J, Gluehmann, M, Janell, D, Bashan, A, Bartels, H, Agmon, I, Franceschi, F, Yonath, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution.
Cell(Cambridge,Mass.), 102, 2000
|
|
4KK0
 
 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
4KK1
 
 | | Crystal Structure of TSC1 core domain from S. pombe | | Descriptor: | Tuberous sclerosis 1 protein homolog | | Authors: | Sun, W, Zhu, Y, Wang, Z.Z, Zhong, Q, Gao, F, Lou, J.Z, Gong, W.M, Xu, W.Q. | | Deposit date: | 2013-05-05 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the yeast TSC1 core domain and implications for tuberous sclerosis pathological mutations.
Nat Commun, 4, 2013
|
|
8G05
 
 | | Cryo-EM structure of an orphan GPCR-Gi protein signaling complex | | Descriptor: | 6-(octylamino)pyrimidine-2,4(3H,5H)-dione, CHOLESTEROL, G-protein coupled receptor 84, ... | | Authors: | Zhang, X, Wang, Y.J, Li, X, Liu, G.B, Gong, W.M, Zhang, C. | | Deposit date: | 2023-01-31 | | Release date: | 2023-11-01 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Pro-phagocytic function and structural basis of GPR84 signaling.
Nat Commun, 14, 2023
|
|
8H7V
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H85
 
 | | Trans-3/4-proline-hydroxylase H11 with 3-hydroxyl-proline | | Descriptor: | 3-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7T
 
 | | Trans-3/4-proline-hydroxylase H11 apo structure | | Descriptor: | CHLORIDE ION, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H81
 
 | | Trans-3/4-proline-hydroxylase H11 with 4-Hydroxyl-proline | | Descriptor: | 4-HYDROXYPROLINE, Phytanoyl-CoA dioxygenase | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H7Y
 
 | | Trans-3/4-proline-hydroxylase H11 with AKG and L-proline | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, PROLINE, ... | | Authors: | Gong, W.M, Hu, X.Y. | | Deposit date: | 2022-10-21 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of L-proline trans-hydroxylase reveal the catalytic specificity and provide deeper insight into AKG-dependent hydroxylation.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8H0O
 
 | | Crystal structure of human serum albumin and ruthenium PZA complex adduct | | Descriptor: | Albumin, CHLORIDE ION, NITRIC OXIDE, ... | | Authors: | Gong, W.J, Wang, Y, Bai, H.H, Wang, W.M, Wang, H.F. | | Deposit date: | 2022-09-30 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.479 Å) | | Cite: | Crystal structure of human serum albumin and ruthenium PZA complex adduct
To Be Published
|
|
8W73
 
 | | Fe-O nanocluster of form-I in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W7V
 
 | | Udif-E164A-E168A soaking in Fe2+ solution for 50 minutes | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W79
 
 | | Fe-O nanocluster of form-III in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
