6AAH
 
 | | Crystal structure of JAK1 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6AAK
 
 | | Crystal structure of JAK3 in complex with peficitinib | | Descriptor: | 4-[[(1S,3R)-5-oxidanyl-2-adamantyl]amino]-1H-pyrrolo[2,3-b]pyridine-5-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Amano, Y. | | Deposit date: | 2018-07-18 | | Release date: | 2018-08-15 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Discovery and structural characterization of peficitinib (ASP015K) as a novel and potent JAK inhibitor
Bioorg. Med. Chem., 26, 2018
|
|
6VMJ
 
 | |
6VMK
 
 | |
1VJ2
 
 | |
1VKM
 
 | |
1VJ1
 
 | |
1VKH
 
 | |
1J6U
 
 | |
1J5S
 
 | |
2VIF
 
 | | Crystal structure of SOCS6 SH2 domain in complex with a c-KIT phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, MAST/STEM CELL GROWTH FACTOR RECEPTOR, SUPPRESSOR OF CYTOKINE SIGNALLING 6 | | Authors: | Bullock, A, Pike, A.C.W, Savitsky, P, Keates, T, Pilka, E.S, von Delft, F, Edwards, A, Weigelt, J, Arrowsmith, C.H, Knapp, S. | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis for C-Kit Inhibition by the Suppressor of Cytokine Signaling 6 (Socs6) Ubiquitin Ligase.
J.Biol.Chem., 286, 2011
|
|
1JNJ
 
 | | NMR solution structure of the human beta2-microglobulin | | Descriptor: | beta2-microglobulin | | Authors: | Verdone, G, Corazza, A, Viglino, P, Pettirossi, F, Giorgetti, S, Mangione, P, Andreola, A, Stoppini, M, Bellotti, V, Esposito, G. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of human beta2-microglobulin reveals the prodromes of its amyloid transition.
Protein Sci., 11, 2002
|
|
3HWN
 
 | | CATHEPSIN L with AZ13010160 | | Descriptor: | Cathepsin L1, Nalpha-[(3-tert-butyl-1-methyl-1H-pyrazol-5-yl)carbonyl]-N-[(2E)-2-iminoethyl]-3-{5-[(Z)-iminomethyl]-1,3,4-oxadiazol-2-yl}-L-phenylalaninamide | | Authors: | Kenny, P, Morley, A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-09-15 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Design of selective Cathepsin inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6G88
 
 | | Crystal structure of Enterococcus Faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | (2R)-2-[(1R)-1-{[(2Z)-2-(5-amino-1,2,4-thiadiazol-3-yl)-2-(hydroxyimino)acetyl]amino}-2-oxoethyl]-5-({2-oxo-1-[(3R)-pyrrolidin-3-yl]-2,5-dihydro-1H-pyrrol-3-yl}methyl)-3,6-dihydro-2H-1,3-thiazine-4-carboxylic acid, Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-04-08 | | Release date: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
6G0K
 
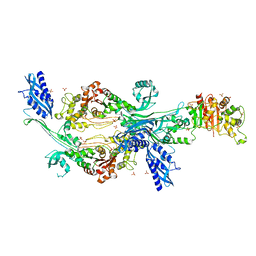 | | Crystal structure of Enterococcus faecium D63r Penicillin-Binding protein 5 (PBP5fm) | | Descriptor: | Low affinity penicillin-binding protein 5 (PBP5), SULFATE ION | | Authors: | Sauvage, E, El Gachi, M, Herman, R, Kerff, F, Charlier, P. | | Deposit date: | 2018-03-19 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of inactivation of Enterococcus faecium penicillin binding protein 5 by ceftobiprole.
To Be Published
|
|
4AMH
 
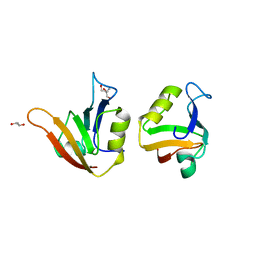 | | Influence of circular permutation on the folding pathway of a PDZ domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DISKS LARGE HOMOLOG 1, GLYCEROL | | Authors: | Hultqvist, G, Punekar, A.S, Chi, C.N, Selmer, M, Gianni, S, Jemth, P. | | Deposit date: | 2012-03-10 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tolerance of Protein Folding to a Circular Permutation in a Pdz Domain
Plos One, 7, 2012
|
|
6FG7
 
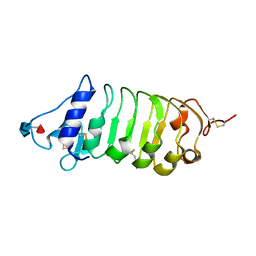 | | Crystal structure of the BIR2 ectodomain from Arabidopsis thaliana. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Inactive LRR receptor-like serine/threonine-protein kinase BIR2, ... | | Authors: | Hothorn, M, Hohmann, U. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling.
Nat Plants, 4, 2018
|
|
6FG8
 
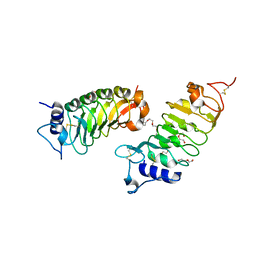 | | Crystal structure of the BIR3 - SERK1 complex from Arabidopsis thaliana. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, PENTAETHYLENE GLYCOL, ... | | Authors: | Hothorn, M, Hohmann, U. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling.
Nat Plants, 4, 2018
|
|
6G3W
 
 | |
4ESR
 
 | | Molecular and Structural Characterization of the SH3 Domain of AHI-1 in Regulation of Cellular Resistance of BCR-ABL+ Chronic Myeloid Leukemia Cells to Tyrosine Kinase Inhibitors | | Descriptor: | DI(HYDROXYETHYL)ETHER, Jouberin | | Authors: | Van Petegem, X.F, Liu, P.X, Lobo, P, Jiang, X. | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Molecular and structural characterization of the SH3 domain of AHI-1 in regulation of cellular resistance of BCR-ABL(+) chronic myeloid leukemia cells to tyrosine kinase inhibitors.
Proteomics, 12, 2012
|
|
5NJS
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 50s time-delay, phased with 1HEW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
2BH0
 
 | | Crystal structure of a SeMet derivative of EXPA from Bacillus subtilis at 2.5 angstrom | | Descriptor: | YOAJ | | Authors: | Petrella, S, Herman, R, Sauvage, E, Filee, P, Joris, B, Charlier, P. | | Deposit date: | 2005-01-06 | | Release date: | 2006-06-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Activity of Bacillus Subtilis Yoaj (Exlx1), a Bacterial Expansin that Promotes Root Colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3T0T
 
 | | Crystal structure of S. aureus Pyruvate Kinase | | Descriptor: | N'-[(1E)-1-(1H-benzimidazol-2-yl)ethylidene]-5-bromo-2-hydroxybenzohydrazide, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cheminformatics-driven discovery of selective, nanomolar inhibitors for staphylococcal pyruvate kinase.
Acs Chem.Biol., 7, 2012
|
|
5X5G
 
 | | Crystal structure of TLA-3 extended-spectrum beta-lactamase in a complex with OP0595 | | Descriptor: | (2S,5R)-N-(2-aminoethoxy)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, SODIUM ION, ... | | Authors: | Wachino, J, Jin, W, Arakawa, Y. | | Deposit date: | 2017-02-15 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the TLA-3 Extended-Spectrum beta-Lactamase and Its Inhibition by Avibactam and OP0595.
Antimicrob. Agents Chemother., 61, 2017
|
|
5NJP
 
 | | Mix-and-diffuse serial synchrotron crystallography: structure of N,N',N''-Triacetylchitotriose bound to Lysozyme with 1s time-delay, phased with 1HEW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Oberthuer, D, Meents, A, Beyerlein, K.R, Chapman, H.N, Lieseke, J. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mix-and-diffuse serial synchrotron crystallography.
IUCrJ, 4, 2017
|
|
