4JGC
 
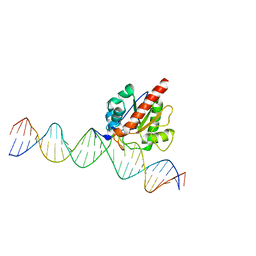 | | Human TDG N140A mutant IN A COMPLEX WITH 5-carboxylcytosine (5caC) | | Descriptor: | 4-amino-2-oxo-1,2-dihydropyrimidine-5-carboxylic acid, G/T mismatch-specific thymine DNA glycosylase, oligonucleotide, ... | | Authors: | Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.582 Å) | | Cite: | Activity and crystal structure of human thymine DNA glycosylase mutant N140A with 5-carboxylcytosine DNA at low pH.
Dna Repair, 12, 2013
|
|
4GZN
 
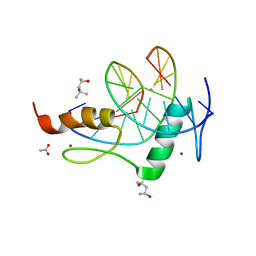 | | Mouse ZFP57 zinc fingers in complex with methylated DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Liu, Y, Zhang, X, Cheng, X. | | Deposit date: | 2012-09-06 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence.
Genes Dev., 26, 2012
|
|
5V3J
 
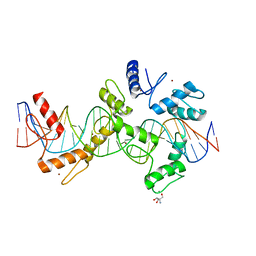 | | mouseZFP568-ZnF1-10 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (26-MER), MAGNESIUM ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
5V3M
 
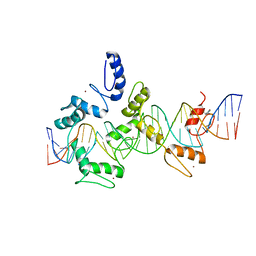 | | mouseZFP568-ZnF1-11 in complex with DNA | | Descriptor: | DNA (28-MER), ZINC ION, Zinc finger protein 568 | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
5UND
 
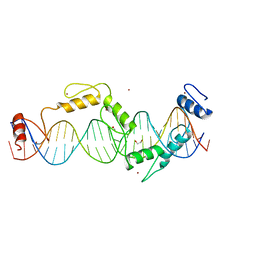 | |
5EYO
 
 | | The crystal structure of the Max bHLH domain in complex with 5-carboxyl cytosine DNA | | Descriptor: | DNA (5'-D(*AP*GP*TP*AP*GP*CP*AP*(1CC)P*GP*TP*GP*CP*TP*AP*CP*T)-3'), Protein max | | Authors: | Wang, D, Hashimoto, H, Zhang, X, Cheng, X. | | Deposit date: | 2015-11-25 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma.
Nucleic Acids Res., 45, 2017
|
|
4R2C
 
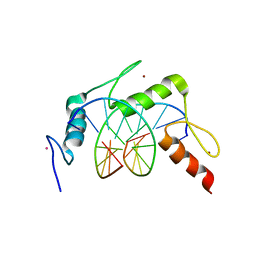 | | Egr1/Zif268 zinc fingers in complex with hydroxymethylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5HC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5HC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Early growth response protein 1, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4M9E
 
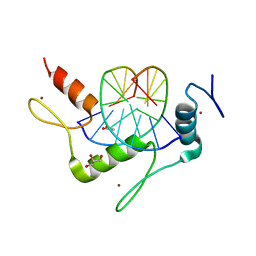 | | Structure of Klf4 zinc finger DNA binding domain in complex with methylated DNA | | Descriptor: | ACETATE ION, DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), ... | | Authors: | Liu, Y, Olanrewaju, Y.O, Blumenthal, R.M, Zhang, X, Cheng, X. | | Deposit date: | 2013-08-14 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for Klf4 recognition of methylated DNA.
Nucleic Acids Res., 42, 2014
|
|
4PBB
 
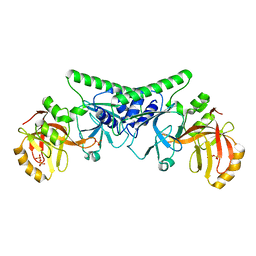 | |
4R2D
 
 | | Egr1/Zif268 zinc fingers in complex with formylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5FC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Early growth response protein 1, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2P
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with hydroxymethylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5HC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5HC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.788 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2Q
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with formylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5FC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5FC)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4RTS
 
 | |
4RTO
 
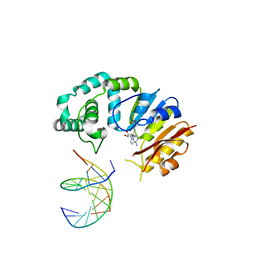 | |
3F8I
 
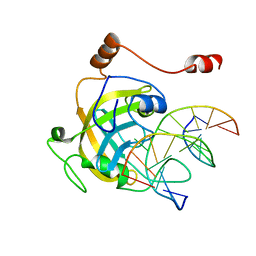 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group P21 | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1 | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
3FDE
 
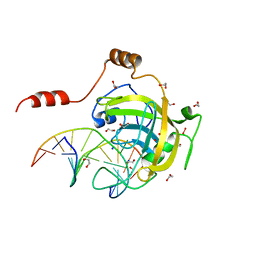 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG DNA, crystal structure in space group C222(1) at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
4LT5
 
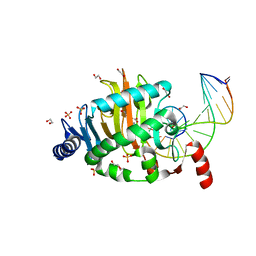 | | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA, MANGANESE (II) ION, ... | | Authors: | Hashimoto, H, Pais, J.E, Zhang, X, Saleh, L, Fu, Z.Q, Dai, N, Correa, I.R, Roberts, R.J, Zheng, Y, Cheng, X. | | Deposit date: | 2013-07-23 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.
Nature, 506, 2014
|
|
4RTR
 
 | |
6BH4
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 5-(1-(tert-butyl)-1H-pyrazol-4-yl)-6-isopropyl-7-oxo-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile (Compound N75/CPI-48) | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-tert-butyl-1H-pyrazol-4-yl)-7-oxo-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH0
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (R)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N51) | | Descriptor: | 2-{(R)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
4R2E
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(5CM)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), Wilms tumor protein, ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-11 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
4R2R
 
 | | Wilms Tumor Protein (WT1) zinc fingers in complex with carboxylated DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*AP*GP*CP*GP*TP*GP*GP*GP*(1CC)P*GP*T)-3'), DNA (5'-D(*TP*AP*(5CM)P*GP*CP*CP*CP*AP*CP*GP*C)-3'), ... | | Authors: | Hashimoto, H, Olanrewaju, Y.O, Zheng, Y, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2014-08-12 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence.
Genes Dev., 28, 2014
|
|
3F8J
 
 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
4PBA
 
 | |
6BGX
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)((4,4-difluorocyclohexyl)methoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N42) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[(4,4-difluorocyclohexyl)methoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
