3C2H
 
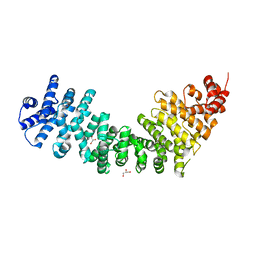 | | Crystal Structure of SYS-1 at 2.6A resolution | | Descriptor: | CITRATE ANION, GLYCEROL, Sys-1 protein | | Authors: | Liu, J, Phillips, B.T, Amaya, M.F, Kimble, J, Xu, W. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The C. elegans SYS-1 protein is a bona fide beta-catenin.
Dev.Cell, 14, 2008
|
|
6TYD
 
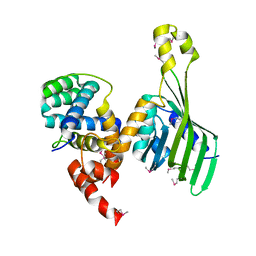 | | Structure of human LDB1 in complex with SSBP2 | | Descriptor: | LIM domain-binding protein 1, Single-stranded DNA-binding protein 2 | | Authors: | Wang, H, Wang, Z, Xu, W. | | Deposit date: | 2019-08-08 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Crystal structure of human LDB1 in complex with SSBP2.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UK5
 
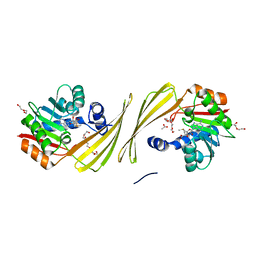 | | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis | | Descriptor: | ACETATE ION, CalS10, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Alvarado, S.K, Miller, M.D, Xu, W, Wang, Z, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of SAM bound CalS10, an amino pentose methyltransferase from Micromonospora echinaspora involved in calicheamicin biosynthesis
To Be Published
|
|
6UBL
 
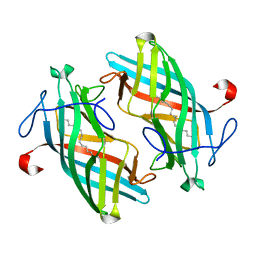 | | Structure of DynF from the Dynemicin Biosynthesis Pathway of Micromonospora chersina | | Descriptor: | DynF, PALMITIC ACID | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Bhardwaj, M, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7N7V
 
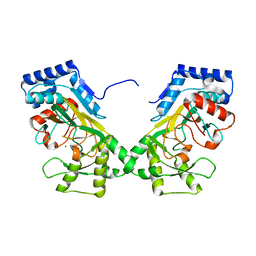 | | Crystal structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes at 2 A. | | Descriptor: | CHLORIDE ION, FE (II) ION, Predicted hydroxylase | | Authors: | Han, L, Xu, W, Ma, M, Miller, M.D, Shen, B, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2021-06-11 | | Release date: | 2022-07-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of TtnM, a Fe(II)-alpha-ketoglutarate-dependent hydroxylase from the tautomycetin biosynthesis pathway in Streptomyces griseochromogenes.
To Be Published
|
|
5WZZ
 
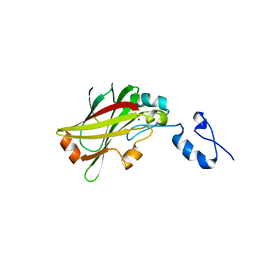 | | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation | | Descriptor: | Axin-1, E3 ubiquitin-protein ligase SIAH1, ZINC ION | | Authors: | Ji, L, Jiang, B, Jiang, X, Charlat, O, Chen, A, Mickanin, C, Bauer, A, Xu, W, Yan, X.-X, Cong, F. | | Deposit date: | 2017-01-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The SIAH E3 ubiquitin ligases promote Wnt/ beta-catenin signaling through mediating Wnt-induced Axin degradation
Genes Dev., 31, 2017
|
|
6PAX
 
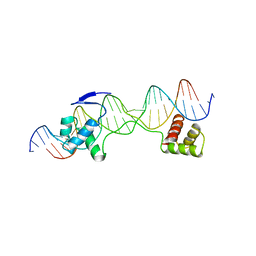 | | CRYSTAL STRUCTURE OF THE HUMAN PAX-6 PAIRED DOMAIN-DNA COMPLEX REVEALS A GENERAL MODEL FOR PAX PROTEIN-DNA INTERACTIONS | | Descriptor: | 26 NUCLEOTIDE DNA, HOMEOBOX PROTEIN PAX-6 | | Authors: | Xu, H.E, Rould, M.A, Xu, W, Epstein, J.A, Maas, R.L, Pabo, C.O. | | Deposit date: | 1999-04-22 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding.
Genes Dev., 13, 1999
|
|
8YM1
 
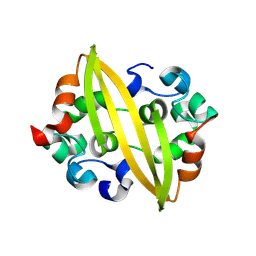 | | Structure of SADS-CoV Virus Nucleocapsid Protein | | Descriptor: | nucleocapsid phosphoprotein | | Authors: | Zhang, Y, Wu, F, Xu, W. | | Deposit date: | 2024-03-08 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unraveling the assembly mechanism of SADS-CoV virus nucleocapsid protein: insights from RNA binding, dimerization, and epitope diversity profiling.
J.Virol., 98, 2024
|
|
3FGA
 
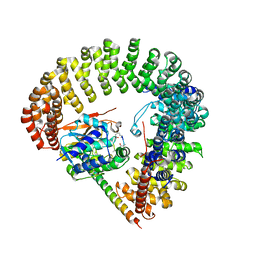 | | Structural Basis of PP2A and Sgo interaction | | Descriptor: | MANGANESE (II) ION, MICROCYSTIN-LR, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Xu, Z, Xu, W. | | Deposit date: | 2008-12-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the PP2A-shugoshin interaction
Mol.Cell, 35, 2009
|
|
7XMN
 
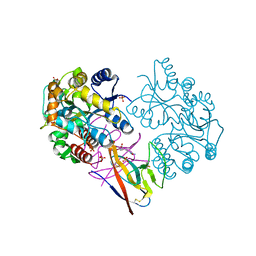 | | Structure of SARS-CoV-2 ORF8 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltodextrin-binding protein, ... | | Authors: | Chen, X, Xu, W. | | Deposit date: | 2022-04-26 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Glycosylated, Lipid-Binding, CDR-Like Domains of SARS-CoV-2 ORF8 Indicate Unique Sites of Immune Regulation.
Microbiol Spectr, 11, 2023
|
|
5JJA
 
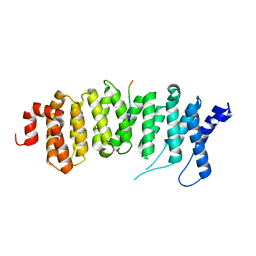 | | Crystal structure of a PP2A B56gamma/BubR1 complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization.
Protein Cell, 7, 2016
|
|
1EXY
 
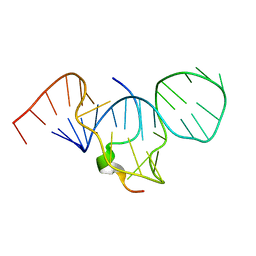 | | SOLUTION STRUCTURE OF HTLV-1 PEPTIDE BOUND TO ITS RNA APTAMER TARGET | | Descriptor: | HTLV-1 REX PEPTIDE, RNA APTAMER, 33-MER | | Authors: | Jiang, F, Gorin, A, Hu, W, Majumdar, A, Baskerville, S, Xu, W, Ellington, A, Patel, D.J. | | Deposit date: | 2000-05-05 | | Release date: | 2000-05-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples.
Structure Fold.Des., 7, 1999
|
|
1G70
 
 | | COMPLEX OF HIV-1 RRE-IIB RNA WITH RSG-1.2 PEPTIDE | | Descriptor: | HIV-1 RRE-IIB 32 NUCLEOTIDE RNA, RSG-1.2 PEPTIDE | | Authors: | Gosser, Y, Hermann, T, Majumdar, A, Hu, W, Frederick, R, Jiang, F, Xu, W, Patel, D.J. | | Deposit date: | 2000-11-08 | | Release date: | 2001-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Peptide-triggered conformational switch in HIV-1 RRE RNA complexes.
Nat.Struct.Biol., 8, 2001
|
|
7MSY
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CALCIUM ION, CHLORIDE ION, CalU17, ... | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
7ML6
 
 | | Structure of CalU17 from the Calicheamicin Biosynthesis Pathway of Micromonospora echinospora | | Descriptor: | CalU17, GLYCEROL | | Authors: | Kosgei, A.J, Miller, M.D, Xu, W, Van Lanen, S.G, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of DynF from the dynemicin-biosynthesis pathway of Micromonospora chersina.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
6XN8
 
 | | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017 | | Descriptor: | 2-hydroxyacyl-CoA lyase 1, 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Miller, M.D, Xu, W, Olmos Jr, J.L, Chou, A, Clomburg, J.M, Gonzalez, R, Philips Jr, G.N. | | Deposit date: | 2020-07-02 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of 2-hydroxyacyl CoA lyase (HACL) from Rhodospirillales bacterium URHD0017
To Be Published
|
|
6XOD
 
 | | Crystal structure of the PEX4-PEX22 protein complex from Arabidopsis thaliana | | Descriptor: | Peroxisome biogenesis protein 22, Protein PEROXIN-4 | | Authors: | Olmos Jr, J.L, Bradford, S.E, Miller, M.D, Xu, W, Wright, Z.J, Bartel, B, Phillips Jr, G.N. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Structure of the Arabidopsis PEX4-PEX22 Peroxin Complex-Insights Into Ubiquitination at the Peroxisomal Membrane
Front Cell Dev Biol, 10, 2022
|
|
6LND
 
 | | Crystal structure of transposition protein TniQ | | Descriptor: | ZINC ION, transposition protein TniQ | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
6LNB
 
 | | CryoEM structure of Cascade-TniQ-dsDNA complex | | Descriptor: | CRISPR RNA (60-MER), CRISPR-associated protein Cas6, CRISPR-associated protein Cas7, ... | | Authors: | Wang, B, Xu, W, Yang, H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of a Tn7-like transposase recruitment and DNA loading to CRISPR-Cas surveillance complex.
Cell Res., 30, 2020
|
|
5CL1
 
 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
1NEM
 
 | | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | | Descriptor: | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXY-D-STREPTAMINE, ... | | Authors: | Jiang, L, Majumdar, A, Hu, W, Jaishree, T.J, Xu, W, Patel, D.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer
Structure Fold.Des., 7, 1999
|
|
1SOI
 
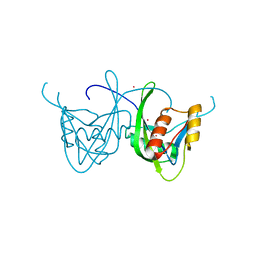 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEX WITH SM+3 | | Descriptor: | MutT/nudix family protein, SAMARIUM (III) ION | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-15 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SZ3
 
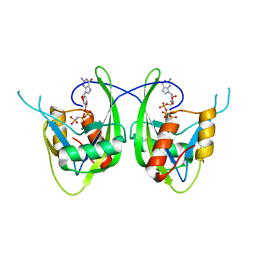 | | CRYSTAL STRUCTURE OF NUDIX HYDROLASE DR1025 IN COMPLEXED WITH GNP AND MG+2 | | Descriptor: | MAGNESIUM ION, MutT/nudix family protein, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-04-02 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
1SJY
 
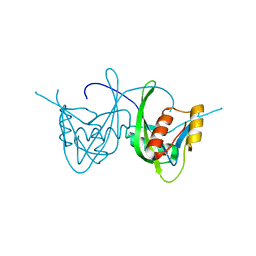 | | Crystal Structure of NUDIX HYDROLASE DR1025 FROM DEINOCOCCUS RADIODURANS | | Descriptor: | MutT/nudix family protein | | Authors: | Ranatunga, W, Hill, E.E, Mooster, J.L, Holbrook, E.L, Schulze-Gahmen, U, Xu, W, Bessman, M.J, Brenner, S.E, Holbrook, S.R, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural Studies of the Nudix Hydrolase DR1025 From Deinococcus radiodurans and its Ligand Complexes.
J.Mol.Biol., 339, 2004
|
|
2Z6H
 
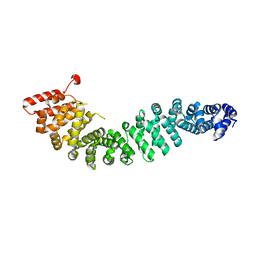 | | Crystal Structure of Beta-Catenin Armadillo Repeat Region and Its C-Terminal domain | | Descriptor: | Catenin beta-1 | | Authors: | Xing, Y, Takemaru, K, Liu, J, Zheng, J, Moon, R, Xu, W. | | Deposit date: | 2007-08-01 | | Release date: | 2008-02-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Full-Length beta-Catenin
Structure, 16, 2008
|
|
