8XFZ
 
 | | The structure of HLA-A/L1-2 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Major capsid protein L1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
4OI7
 
 | | RAGE recognizes nucleic acids and promotes inflammatory responses to DNA | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*AP*CP*GP*TP*TP*CP*GP*TP*AP*GP*CP*AP*TP*CP*GP*TP*TP*GP*CP*AP*G)-3', 5'-D(*CP*TP*GP*CP*AP*AP*CP*GP*AP*TP*GP*CP*TP*AP*CP*GP*AP*AP*CP*GP*TP*G)-3', ... | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.104 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
4OI8
 
 | | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA. | | Descriptor: | 5'-D(*CP*CP*AP*TP*GP*AP*CP*TP*GP*TP*AP*GP*GP*AP*AP*AP*CP*TP*CP*TP*AP*GP*A)-3', 5'-D(*CP*TP*CP*TP*AP*GP*AP*GP*TP*TP*TP*CP*CP*TP*AP*CP*AP*GP*TP*CP*AP*TP*G)-3', Advanced glycosylation end product-specific receptor | | Authors: | Jin, T, Jiang, J, Xiao, T. | | Deposit date: | 2014-01-19 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA.
J.Exp.Med., 210, 2013
|
|
4F58
 
 | |
8UJY
 
 | | Crystal structure of human WD repeat-containing protein 5 in complex with 4-(3,5-dimethoxybenzyl)-9-(4-fluoro-2-methylphenyl)-7-((2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl)methyl)-3,4-dihydrobenzo[f][1,4]oxazepin-5(2H)-one (compound 8) | | Descriptor: | (9P)-4-[(3,5-dimethoxyphenyl)methyl]-9-(4-fluoro-2-methylphenyl)-7-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-3,4-dihydro-1,4-benzoxazepin-5(2H)-one, WD repeat-containing protein 5 | | Authors: | Zhao, B, Amporndanai, K, Fesik, S.W. | | Deposit date: | 2023-10-11 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-Based Discovery of Potent, Orally Bioavailable Benzoxazepinone-Based WD Repeat Domain 5 Inhibitors.
J.Med.Chem., 66, 2023
|
|
4F57
 
 | |
7U9Y
 
 | |
7UAS
 
 | |
8GRQ
 
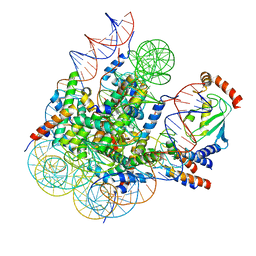 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | Descriptor: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GRM
 
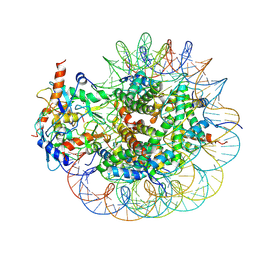 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | Descriptor: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
7E23
 
 | | SARS-CoV-2 spike in complex with the CA521 neutralizing antibody Fab (focused refinement on Fab-RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CA521 Heavy Chain, CA521 Light Chain, ... | | Authors: | Liu, C, Song, D, Dou, C. | | Deposit date: | 2021-02-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and function analysis of a potent human neutralizing antibody CA521 FALA against SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WM1
 
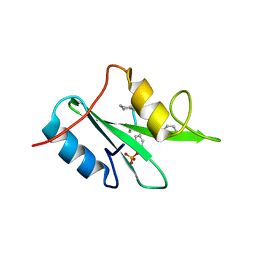 | |
6WO2
 
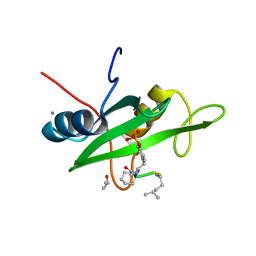 | |
2ZUW
 
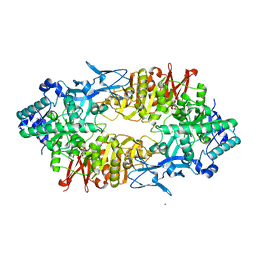 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUT
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUS
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase | | Descriptor: | Lacto-N-biose phosphorylase, MAGNESIUM ION | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUV
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc, Ethylene glycol, and nitrate | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
2ZUU
 
 | | Crystal structure of Galacto-N-biose/Lacto-N-biose I phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, GLYCEROL, Lacto-N-biose phosphorylase, ... | | Authors: | Hidaka, M, Nishimoto, M, Kitaoka, M, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2008-10-28 | | Release date: | 2008-12-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: A large deformation of a tim barrel scaffold
J.Biol.Chem., 284, 2009
|
|
7ZRB
 
 | |
