1QVJ
 
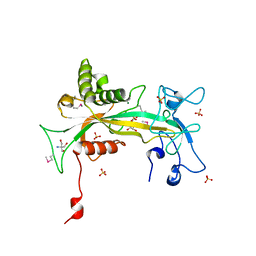 | | structure of NUDT9 complexed with ribose-5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-beta-D-ribofuranose, ADP-ribose pyrophosphatase, ... | | Authors: | Shen, B.W, Perraud, A.-L, Scharenberg, A.S, Stoddard, B.L. | | Deposit date: | 2003-08-27 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The crystal structure and mutational analysis of human NUDT9
J.Mol.Biol., 332, 2003
|
|
301D
 
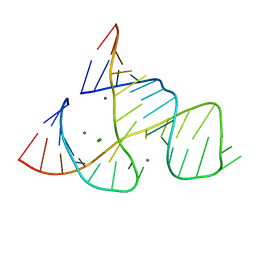 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MG(II)-SOAKED | | Descriptor: | MAGNESIUM ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
300D
 
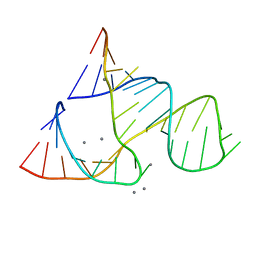 | | CAPTURING THE STRUCTURE OF A CATALYTIC RNA INTERMEDIATE: RNA HAMMERHEAD RIBOZYME, MN(II)-SOAKED | | Descriptor: | MANGANESE (II) ION, RNA HAMMERHEAD RIBOZYME | | Authors: | Scott, W.G, Murray, J.B, Arnold, J.R.P, Stoddard, B.L, Klug, A. | | Deposit date: | 1996-12-14 | | Release date: | 1997-01-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme.
Science, 274, 1996
|
|
3BVQ
 
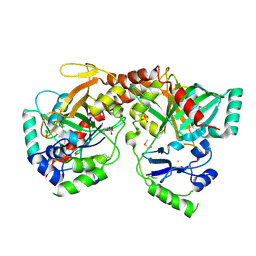 | | Crystal Structure of Apo NotI Restriction Endonuclease | | Descriptor: | FE (III) ION, NotI restriction endonuclease, SULFATE ION | | Authors: | Lambert, A.R, Sussman, D, Shen, B, Stoddard, B.L. | | Deposit date: | 2008-01-07 | | Release date: | 2008-01-22 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of the Rare-Cutting Restriction Endonuclease NotI Reveal a Unique Metal Binding Fold Involved in DNA Binding.
Structure, 16, 2008
|
|
3C25
 
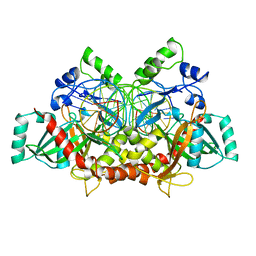 | | Crystal Structure of NotI Restriction Endonuclease Bound to Cognate DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DGP*DGP*DAP*DGP*DGP*DCP*DGP*DCP*DGP*DGP*DCP*DCP*DGP*DCP*DGP*DCP*DCP*DGP*DCP*DCP*DG)-3'), DNA (5'-D(*DCP*DGP*DGP*DCP*DGP*DGP*DCP*DGP*DCP*DGP*DGP*DCP*DCP*DGP*DCP*DGP*DCP*DCP*DTP*DCP*DCP*DG)-3'), ... | | Authors: | Lambert, A.R, Sussman, D, Shen, B, Stoddard, B.L. | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the Rare-Cutting Restriction Endonuclease NotI Reveal a Unique Metal Binding Fold Involved in DNA Binding.
Structure, 16, 2008
|
|
3UGM
 
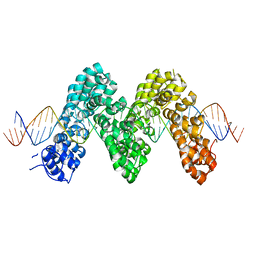 | | Structure of TAL effector PthXo1 bound to its DNA target | | Descriptor: | DNA-1, DNA-2, TAL effector AvrBs3/PthA | | Authors: | Mak, A.N.S, Bradley, P, Cernadas, R.A, Bogdanove, A.J, Stoddard, B.L. | | Deposit date: | 2011-11-02 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of TAL Effector PthXo1 Bound to Its DNA Target.
Science, 335, 2012
|
|
7SQ3
 
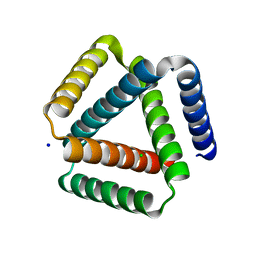 | | Designed trefoil knot protein, variant 1 | | Descriptor: | CHLORIDE ION, Designed trefoil knot protein, variant 1, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.453 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ4
 
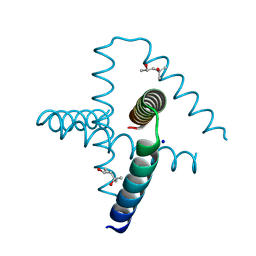 | | Designed trefoil knot protein, variant 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Designed trefoil knot protein, variant 2, ... | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.493 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
7SQ5
 
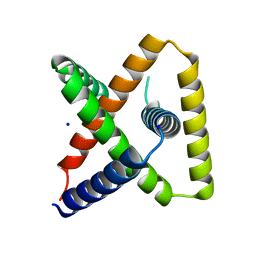 | | Designed trefoil knot protein, variant 3 | | Descriptor: | Designed trefoil knot protein, variant 3, SODIUM ION | | Authors: | Takushi, B, Doyle, L, Stoddard, B.L, Bradley, P. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | De novo design of knotted tandem repeat proteins.
Nat Commun, 14, 2023
|
|
2O6M
 
 | | H98Q mutant of the homing endonuclease I-PPOI complexed with DNA | | Descriptor: | 5'-D(*DTP*DTP*DGP*DAP*DCP*DTP*DCP*DTP*DCP*DTP*DTP*DAP*DAP*DGP*DAP*DGP*DAP*DGP*DTP*DCP*DA)-3', Intron-encoded endonuclease I-PpoI, MAGNESIUM ION, ... | | Authors: | Eastberg, J.H, Stoddard, B.L. | | Deposit date: | 2006-12-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism.
Biochemistry, 46, 2007
|
|
2O3K
 
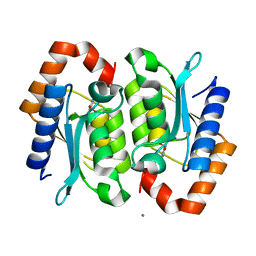 | |
2OST
 
 | | The structure of a bacterial homing endonuclease : I-Ssp6803I | | Descriptor: | CALCIUM ION, Putative endonuclease, Synthetic DNA 29 MER | | Authors: | Zhao, L, Bonocora, R.P, Shub, D.A, Stoddard, B.L. | | Deposit date: | 2007-02-06 | | Release date: | 2007-03-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif.
Embo J., 26, 2007
|
|
6CZI
 
 | | Structure of a redesigned beta barrel, mFAP1, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP1 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6CZG
 
 | |
6CZJ
 
 | |
6CZH
 
 | | Structure of a redesigned beta barrel, mFAP0, bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, mFAP0 | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6D0T
 
 | | De novo design of a fluorescence-activating beta barrel - BB1 | | Descriptor: | BB1 | | Authors: | Dou, J, Vorobieva, A.A, Sheffler, W, Doyle, L.A, Park, H, Bick, M.J, Mao, B, Foight, G.W, Lee, M, Carter, L, Sankaran, B, Ovchinnikov, S, Marcos, E, Huang, P, Vaughan, J.C, Stoddard, B.L, Baker, D. | | Deposit date: | 2018-04-10 | | Release date: | 2018-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | De novo design of a fluorescence-activating beta-barrel.
Nature, 561, 2018
|
|
6MAF
 
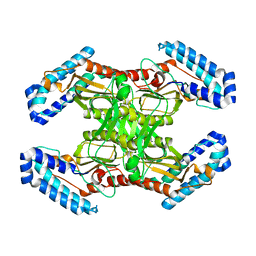 | | native BbvCI A2B2 tetramer at low resolution | | Descriptor: | BbvCI endonuclease subunit 1, BbvCI endonuclease subunit 2 | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Nucleic Acids Res., 47, 2019
|
|
6M9G
 
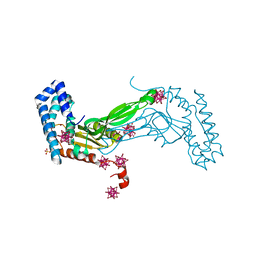 | | BbvCI B2 dimer with Ta6Br14 clusters | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BROMIDE ION, ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2018-08-23 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Nucleic Acids Res., 47, 2019
|
|
6MAG
 
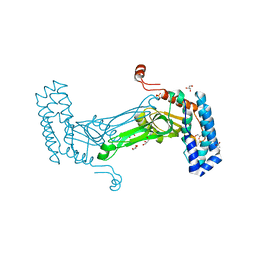 | | native BbvCI B2 dimer in space group C222 | | Descriptor: | ACETATE ION, BbvCI endonuclease subunit 2, GLYCEROL, ... | | Authors: | Shen, B.W, Stoddard, B.L. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure, subunit organization and behavior of the asymmetric Type IIT restriction endonuclease BbvCI.
Nucleic Acids Res., 47, 2019
|
|
6OHH
 
 | | Structure of EF1p2_mFAP2b bound to DFHBI | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, CALCIUM ION, EF1p2_mFAP2b | | Authors: | Doyle, L.A, Stoddard, B.L. | | Deposit date: | 2019-04-05 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of sensing modalities into de novo designed fluorescence-activating proteins
Nat Commun, 12, 2021
|
|
1IPP
 
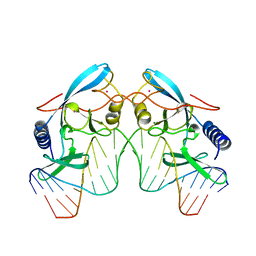 | | HOMING ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CADMIUM ION, DNA (5'-D(*TP*TP*GP*AP*CP*TP*CP*TP*CP*TP*TP*AP*AP*GP*AP*GP*AP*GP*TP*CP*A)-3'), INTRON-ENCODED ENDONUCLEASE I-PPOI, ... | | Authors: | Flick, K.E, Jurica, M.S, Monnat Jr, R.J, Stoddard, B.L. | | Deposit date: | 1998-03-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI.
Nature, 394, 1998
|
|
6UVW
 
 | |
6UWK
 
 | |
6UWJ
 
 | |
