8OVR
 
 | | Clostridium perfringens chitinase CP56_3454 apo form | | Descriptor: | Chitinase B, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), SODIUM ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Clostridium perfringens chitinases, key enzymes during early stages of necrotic enteritis in broiler chickens.
Plos Pathog., 20, 2024
|
|
8OWF
 
 | | Clostridium perfringens chitinase CP4_3455 with chitosan | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Clostridium perfringens chitinases, key enzymes during early stages of necrotic enteritis in broiler chickens.
Plos Pathog., 20, 2024
|
|
8OXU
 
 | | Crystal Structure of the Hsp90-LA1011 Complex | | Descriptor: | ATP-dependent molecular chaperone HSP82, dimethyl 2,6-bis[2-(dimethylamino)ethyl]-1-methyl-4-[4-(trifluoromethyl)phenyl]-4~{H}-pyridine-3,5-dicarboxylate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2023-05-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer's Disease.
Biomolecules, 13, 2023
|
|
8PFF
 
 | | Galectin-3C in complex with a triazolesulfone derivative | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-(phenylsulfonyl)-1,2,3-triazol-1-yl]oxan-2-yl]sulfanyl-oxane-3,4,5-triol, Galectin-3, MAGNESIUM ION, ... | | Authors: | Kumar, R, Mahanti, M, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2023-06-15 | | Release date: | 2023-11-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ligand Sulfur Oxidation State Progressively Alters Galectin-3-Ligand Complex Conformations To Induce Affinity-Influencing Hydrogen Bonds.
J.Med.Chem., 66, 2023
|
|
9BCM
 
 | |
9BEL
 
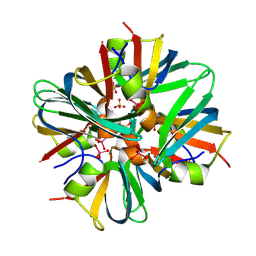 | | Tungstate binding protein (Tungbindin) from Eubacterium limosum with five Tungstates bound | | Descriptor: | Molybdenum-pterin binding domain-containing protein, SULFATE ION, TUNGSTATE(VI)ION | | Authors: | Zhou, D, Rose, J.P, Chen, L, Wang, B.C. | | Deposit date: | 2024-04-15 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
9BA2
 
 | | Crystal structure of the binary complex of DCAF1 and WDR5 | | Descriptor: | DDB1- and CUL4-associated factor 1, IMIDAZOLE, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Srivastava, S, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
9BD5
 
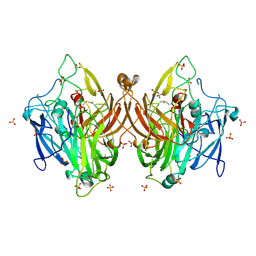 | | Laccase from Bacillus licheniformis | | Descriptor: | COPPER (II) ION, SULFATE ION, Spore coat protein A | | Authors: | Habib, M.H, Smith, T.J. | | Deposit date: | 2024-04-11 | | Release date: | 2024-11-20 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Polymerization potential of a bacterial CotA-laccase for beta-naphthol: enzyme structure and comprehensive polymer characterization.
Front Microbiol, 15, 2024
|
|
9B8J
 
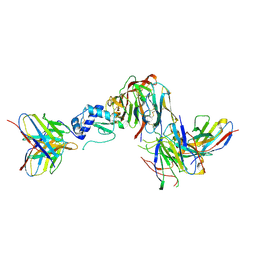 | | GP38-GnH-DS-Gc in the pre-fusion conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADI-36125 heavy chain, ADI-36125 light chain, ... | | Authors: | McFadden, E, McLellan, J.S. | | Deposit date: | 2024-03-30 | | Release date: | 2024-12-11 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Engineering and structures of Crimean-Congo hemorrhagic fever virus glycoprotein complexes.
Cell, 188, 2025
|
|
8XWX
 
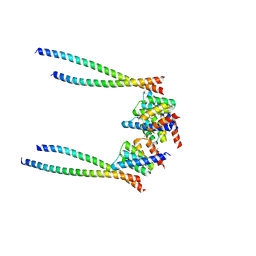 | |
9ARN
 
 | |
9ARP
 
 | |
9AZB
 
 | | Crystal structure of LolTv5 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-11 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
9AZA
 
 | | Crystal structure of LolTv4 | | Descriptor: | Aminotransferase, class V/Cysteine desulfurase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Gao, J, Hai, Y. | | Deposit date: | 2024-03-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Enzymatic Synthesis of Unprotected alpha , beta-Diamino Acids via Direct Asymmetric Mannich Reactions.
J.Am.Chem.Soc., 146, 2024
|
|
1VKA
 
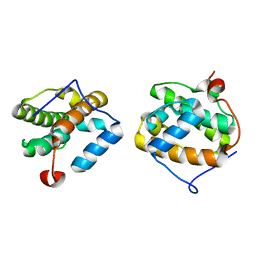 | | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment | | Descriptor: | Microtubule-associated protein RP/EB family member 1 | | Authors: | Liu, Z.-J, Tempel, W, Schubot, F.D, Shah, A, Dailey, T.A, Mayer, M.R, Rose, J.P, Dailey, H.A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-10 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Southeast Collaboratory for Structural Genomics: Hypothetical Human Protein Q15691 N-Terminal Fragment
TO BE PUBLISHED
|
|
5MJL
 
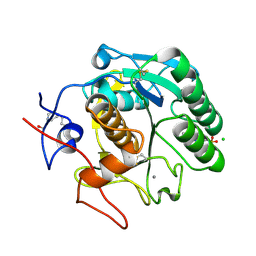 | | Single-shot pink beam serial crystallography: Proteinase K | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21013784 Å) | | Cite: | Pink-beam serial crystallography.
Nat Commun, 8, 2017
|
|
5MJG
 
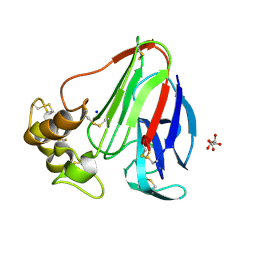 | | Single-shot pink beam serial crystallography: Thaumatin | | Descriptor: | S,R MESO-TARTARIC ACID, SODIUM ION, Thaumatin-1 | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-shot pink beam serial crystallography: Thaumatin
To Be Published
|
|
5ZYR
 
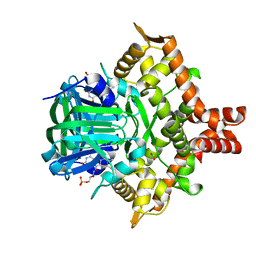 | | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, ... | | Authors: | Oonanant, W, Phongsak, T, Sucharitakul, J, Chaiyen, P, Yuvaniyama, J. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.20001316 Å) | | Cite: | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii
To Be Published
|
|
7S1G
 
 | | wild-type Escherichia coli stalled ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1H
 
 | | Wild-type Escherichia coli ribosome with antibiotic linezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S86
 
 | |
7S7S
 
 | |
4YE7
 
 | |
6RPQ
 
 | | Crystal structure of PhoCDC21-1 intein | | Descriptor: | Ubiquitin-like protein SMT3,1108aa long hypothetical cell division control protein | | Authors: | Beyer, H.M, Mikula, K.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
6RPP
 
 | | Crystal structure of PabCDC21-1 intein | | Descriptor: | ACETATE ION, Cell division control protein, DI(HYDROXYETHYL)ETHER | | Authors: | Mikula, K.M, Beyer, H.M, Iwai, H. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of CDC21-1 inteins from hyperthermophilic archaea reveal the selection mechanism for the highly conserved homing endonuclease insertion site.
Extremophiles, 23, 2019
|
|
